According to a recent study of information from the Veterans Affairs Million Veteran Program, there are genetic correlations between COVID-19 severity and specific medical disorders that are established risk factors for severe COVID-19. The findings were published in the open-access journal PLOS Genetics on April 28th,2022, by Anurag Verma of the Corporal Michael Crescenz VA Medical Center in Philadelphia, Pennsylvania, United States, and colleagues.
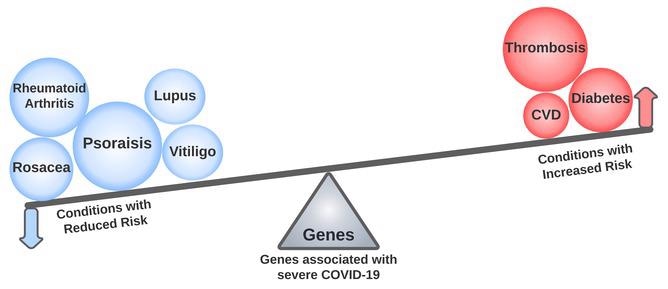 While genes linked to severe COVID-19 were associated with established risk factors and adverse outcomes, including deep vein thrombosis, a significant subset of these genes had opposite associations with reduced risk of immune-mediated disorders such as psoriasis, lupus, and rheumatoid arthritis. Image Credit: Anurag Verma, Katherine Liao, and Scott Damrauer (CC-BY 4.0, https://creativecommons.org/licenses/by/4.0/).
While genes linked to severe COVID-19 were associated with established risk factors and adverse outcomes, including deep vein thrombosis, a significant subset of these genes had opposite associations with reduced risk of immune-mediated disorders such as psoriasis, lupus, and rheumatoid arthritis. Image Credit: Anurag Verma, Katherine Liao, and Scott Damrauer (CC-BY 4.0, https://creativecommons.org/licenses/by/4.0/).
Some patients with COVID-19 have a more severe case of the disease than others. A previous study has linked some human gene variants to a person who has COVID-19 that is more severe. Some of these variants may be linked to other medical disorders that are currently well established; discovering these shared variants might help researchers better understand COVID-19 and open up new therapeutic options.
Verma and colleagues used an unparalleled collection of genotypic networks connected to electronic health record data (EHR) for over 650,000 U.S. veterans to find shared variants. Researchers used a technique called a phenome-wide association study (PheWAS) to explore correlations between polymorphisms often seen in Veterans with severe COVID-19 and variants linked to a variety of other disorders.
Certain COVID-19-associated variations were found to be linked to known COVID-19 risk factors, according to the study. Variants linked to venous embolism and thrombosis, as also type 2 diabetes and ischemic heart disease—two recognized COVID-19 risk factors—were shown to have particularly significant links.
For Veterans of African and Hispanic origin, genetic correlations between severe COVID-19 and neutropenia were discovered, but not for those of European ancestry.
Idiopathic pulmonary fibrosis and persistent alveolar lung disease were linked to severe COVID-19 genetically, while some other respiratory infections, as well as chronic obstructive pulmonary disease (COPD), were not.
Some of the variations linked to severe COVID-19 were also linked to a lower incidence of autoimmune diseases including psoriasis and lupus. When creating novel therapies, these findings underscore the need to carefully weigh diverse parts of the immune system.
Despite the PheWAS method’s shortcomings, these findings might help researchers better understand COVID-19 and lead to the development of novel therapies.
The study demonstrates the value and impact of large biobanks linking genetic variations with EHR data in public health response to the current and future pandemics. MVP is one of the most diverse cohorts in the US. We had a unique opportunity to scan thousands of conditions documented before the COVID-19 pandemic. We gained insights into the genetic architecture of COVID-19 risk factors and disease complication.”
Anurag Verma, Corporal Michael Crescenz VA Medical Center
“One thing that stood out to us was the high number of immune-mediated conditions that shared genetic architecture with severe manifestations of COVID-19. The nature of the associations brought to light how the SARS-CoV2 virus pushes on a pressure point in the human immune system and its constant balancing act of fighting infection while maintaining enough control so that it does not also become an autoimmune process, attacking self.” coauthor Katherine Liao concludes.
Source:
Journal reference:
Verma, A., et al. (2022) A Phenome-Wide Association Study of genes associated with COVID-19 severity reveals shared genetics with complex diseases in the Million Veteran Program. PLOS Genetics. doi.org/10.1371/journal.pgen.1010113.