According to a recent Duke research in mice, the microbes that assist break down food instruct the gut on how to do their job better.
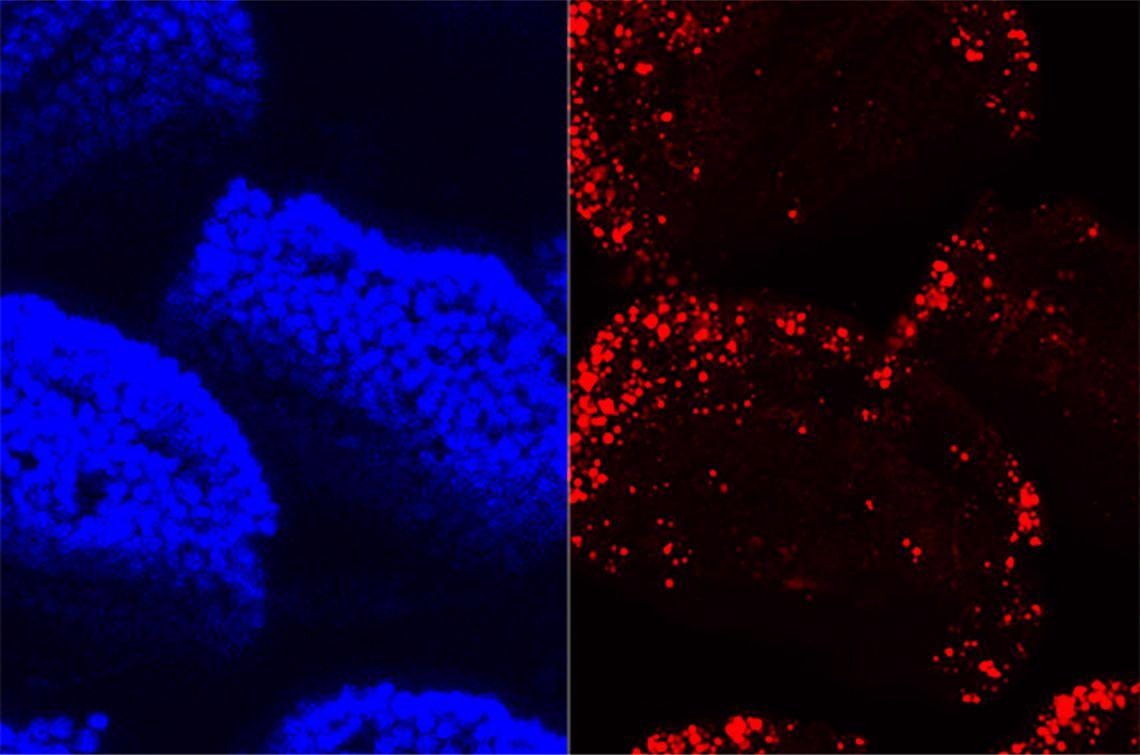 A confocal microscopy image of villi in the small intestine shows cell nuclei in blue and absorbed dietary fat in red. Image Credit: John Rawls Lab.
A confocal microscopy image of villi in the small intestine shows cell nuclei in blue and absorbed dietary fat in red. Image Credit: John Rawls Lab.
According to the researchers, microbes appear to have the ability to influence which genes in the gut are activated, termed an action, in turn, so that interaction may lead to a remodeling of the gut’s epithelial cells lining the gut to fit the diet.
The gut is a fascinating interface between an animal and the world it lives in, and it receives information from both the diet and the microbes it harbors.”
John Rawls, PhD, Professor and Director, Molecular Genomics and Microbiology, Duke Microbiome Center
Cellular and Molecular Gastroenterology and Hepatology, an open-access journal, published the research on May 6th, 2022.
The Duke researchers compared mice raised without gut microbes to mice grown with a normal gut microbiome to begin parsing the information flowing from the microbes to the gut cells. The researchers looked at the interactions between RNA transcription (DNA being copied to RNA) and the proteins that switch this replicating process on or off in the small intestine, which is where the majority of fat and other nutrient absorption takes place.
While both germ-free and normal mice were able to ingest fatty acids in a high-fat diet, the germ-free animals employed a whole distinct collection of genes to deal with the high-fat meal.
We were surprised to find that the gene playbook that the gut epithelium uses to respond to dietary fat is different depending on whether or not microbes are there.”
John Rawls, PhD, Professor and Director, Molecular Genomics and Microbiology, Duke Microbiome Center
The researchers also discovered that microbes can enhance fat absorption in the gut.
It’s a relatively consistent finding across multiple studies, from our lab and others, that microbes actually promote lipid absorption. And that, at some level, also impacts systemic processes like weight gain.”
Colin Lickwar, PhD, Study First Author and Senior Research Associate, Rawls’ lab, Duke University
The activity of genes associated with fatty acid oxidation, or the burning of fatty acids to generate fuel for the gut’s cells, increased in germ-free mice.
Rawls noted, “Typically we think about the gut just doing its job absorbing dietary nutrients across the epithelium to share with the rest of the body, but the gut has to eat too. So what we think is going on in germ-free animals, is that the gut is consuming more of the fat than it would if the microbes were there.”
This might be due to changes in the gut epithelial cell composition.
Lickwar explained, “There are a bunch of recent papers showing that there is a substantial capacity to change the larger architecture of the intestine as well as in the individual gene programs. There is a remarkable amount of plasticity in the intestine. We largely don’t understand it, but some of it is elucidated by this paper.”
The researchers concentrated their efforts on HNF4-Alpha, a transcription factor that is known to control genes associated with lipid metabolism and microbial response.
“We thought that it might represent an interface or a crossroads between interpreting information that comes from either microbial sources or from dietary fat. It’s certainly complicated, but we do appear to identify that HNF4-Alpha is important in simultaneously integrating multiple signals within the intestine,” Lickwar added.
“For every way that germ-free animals seem unusual, that teaches us something about the large impact of the microbiome on what we consider to be ‘normal’ animal biology,” Rawls concluded.
Source:
Journal reference:
Lickwar, C. R., et al. (2022) Transcriptional integration of distinct microbial and nutritional signals by the small intestinal epithelium. Cellular and Molecular Gastroenterology and Hepatology. doi.org/10.1016/j.jcmgh.2022.04.013.