Tuberculosis is a stubborn disease caused by even more stubborn bacteria. While many bacterial infections clear up within days of taking antibiotics, TB can take up to six months to clear up and, in some cases, never leave the human body. In 2020, it claimed 1.5 million lives, second only to COVID-19 in terms of infectious disease deaths.
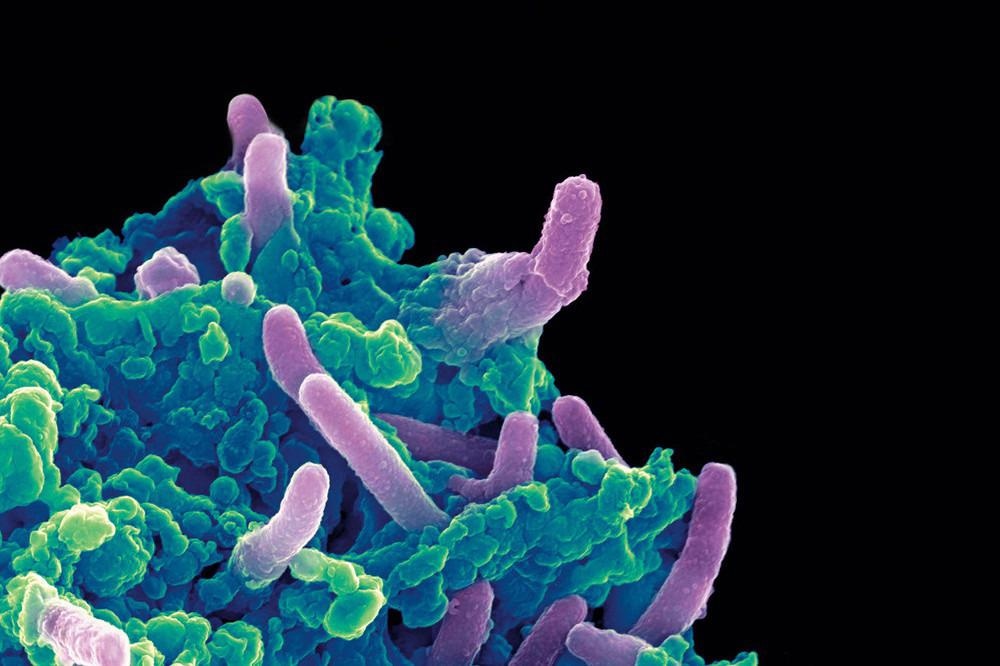 Colored scanning electron micrograph of Mycobacterium tuberculosis bacteria (purple) infecting a macrophage. Image Credit: The Rockefeller University.
Colored scanning electron micrograph of Mycobacterium tuberculosis bacteria (purple) infecting a macrophage. Image Credit: The Rockefeller University.
A new study published in Nature Microbiology outlines the mechanisms by which Mycobacterium tuberculosis (Mtb) bacteria resist antibiotics, revealing hundreds of drug targets that might help Mtb lose its infamous resistance. The researchers also discovered a class of current antibiotics that might be particularly effective against one of Southeast Asia’s most common strains.
Examining how current drugs affect bacterial physiology, and how the bacterium subverts this, is part of our long-term goal of developing better treatment combinations. This study is the tip of the spear getting us into that realm.”
Jeremy Rock, Head, Laboratory of Host-Pathogen Biology, The Rockefeller University
A genome-wide view of Mtb
The researchers started with a simple question.
We just wanted to know all of the genes involved in Mtb’s resistance to different antibiotics.”
Nick Poulton, Study Co-Author and Graduate Student, Laboratory of Host-Pathogen Biology, The Rockefeller University
The researchers created a platform based on the CRISPR gene knockdown technique that scanned the Mtb genome and pitted the bacteria against popular antibiotics to see how the presence or absence of each gene affected the efficacy of current drugs.
“This allowed us to study essentially every gene in the Mtb genome in a single pooled library, and we were able to inhibit the expression of specific genes without making permanent changes to the bacterial genome,” Poulton adds.
Researchers eventually found 1,373 genes that, when silenced, made Mtb susceptible to antibiotics, as well as 775 genes that had the reverse effect—when the latter genes were stopped, the bacteria became more resistant to medications.
Mtb was highly susceptible to current therapies due to two genes, in particular, mtrA and mtrB. These genes are in charge of keeping the bacterium’s protective covering in place, and they are thought to play a role in Mtb’s natural resistance to antibiotics. Future therapies, the researchers believe, will improve the effectiveness of present antibiotics by disrupting the genes’ activity.
When you inhibit these genes, a number of bacterial-cell processes goes away. This renders Mtb sensitive to many drugs that would otherwise have been less effective.”
Shuqi Li, Study Co-Author and Former Postdoctoral Fellow, Laboratory of Host-Pathogen Biology, The Rockefeller University
In addition, Rock, Poulton, Li, and colleagues discovered several previously unknown pathways by which mutations in the bacA gene lead to drug resistance.
Poulton states, “We’re seeing treatment failure all over the place, and we wanted to better understand the genetic mechanisms that may be leading to resistance. The bacA mutations that we discovered may be an unappreciated source for resistance to second-line drugs, and should be monitored more closely in the clinic.”
Optimizing new antibiotics
Following these preliminary results, the researchers focused on linezolid, a recently authorized antibiotic that is very effective against Mtb but has substantial adverse effects. Rock and colleagues wondered if their findings may help guide ongoing work throughout the world to improve linezolid so that it is still effective at lower, less toxic dosages.
Indeed, researchers discovered two genes that, when blocked, made Mtb more susceptible to linezolid by working on ribosomes, the molecular engines that convert RNA into protein in bacteria. Because linezolid works by gumming up bacterial ribosomes, the scientists claim that these genes fight back against the antibiotic by restarting frozen protein machinery, contributing to resistance.
Non-resistant Mtb became twelve times more vulnerable to linezolid after the researchers silenced both genes. Even more surprising were their findings in drug-resistant Mtb: knocking out both genes appeared to completely reverse antibiotic resistance, converting resistant Mtb back into normal Mtb. “If you could find drugs that inhibit these two pathways, it might theoretically be possible to fully restore linezolid susceptibility back to wild type levels,” Poulton explains.
Inhibiting those two genes, even in non-resistant strains, might increase linezolid effectiveness sufficiently to allow physicians to use lower dosages, potentially minimizing negative side effects.
Dusting off shelved drugs
These discoveries might have far-reaching implications for drug makers attempting to combat the rising problem of antibiotic-resistant tuberculosis in the future. In addition, the Rock lab-produced another finding that might save lives in the short term.
The researchers discovered that one Mtb strain, which is responsible for half a million tuberculosis infections in Southeast Asia each year, had spontaneously acquired an unusual mutation in the whiB7 gene.
Scientists believe the mutation occurred some 900 years ago, but it is unclear what benefit, if any, it provided. What is obvious is that this mutation makes the bacteria very sensitive to a family of antibiotics known as macrolides, which are safe, well-tolerated, and FDA-approved.
Although macrolides are commonly recommended to treat other infections, they have never been used to treat tuberculosis.
“Our research in vitro and in mice suggests that this lineage may be hyper-susceptible to macrolide drugs,” Rock concludes.
Source:
Journal reference:
Li, S., et al. (2022) CRISPRi chemical genetics and comparative genomics identify genes mediating drug potency in Mycobacterium tuberculosis. Nature Microbiology. doi.org/10.1038/s41564-022-01130-y.