Highly structured chromatin encodes the program of life with only one set of genes in the core of every cell. Since various genes are stimulated at different periods of life while staying silent elsewhere, this is feasible. Some genes from a former life stage are hidden deep within the nuclear perimeter, while others manage to break out.
 Aberrant activation of developmentally restricted genes caused by large-scale epigenome remodeling is a driver of human stem cell aging. Image Credit: Liu et al.
Aberrant activation of developmentally restricted genes caused by large-scale epigenome remodeling is a driver of human stem cell aging. Image Credit: Liu et al.
Scientists have been able to figure out how this hierarchical system keeps everything in line. However, according to a new study, the system becomes chaotic as people age. Chaos emerges particularly from the nuclear periphery, leading to large-scale chromatin reconfiguration and epigenetic instability, which eventually leads to cellular senescence.
Scientists from the Institute of Zoology and the Beijing Institute of Genomics of the Chinese Academy of Sciences partnered in a recent work published in Developmental Cell to unravel the large-scale modification of the epigenomic landscape throughout aging. They discovered that abnormal placental-related gene expression is a significant driver and molecular biomarker of cellular aging.
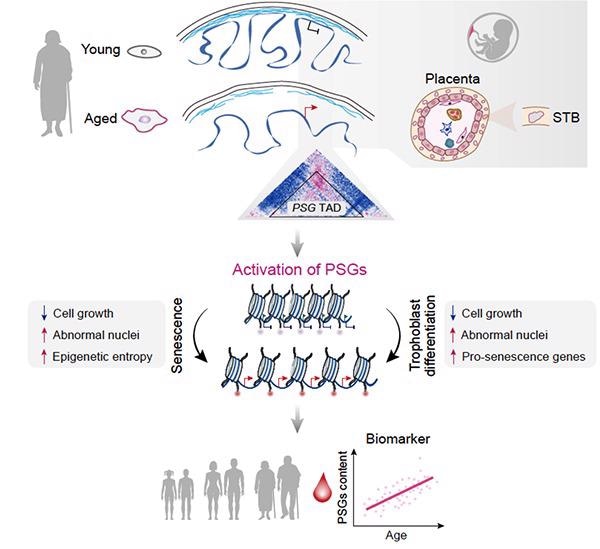
Aging is linked to stem cell exhaustion, which is characterized by a wide range of epigenetic abnormalities. At the cellular level, malformed nuclei are common in elderly human stem cells. However, it was unclear how this aging phenotype connected to the aged epigenetic state.
The scientists created a large-scale, high-resolution epigenomic landscape resource using isogenic juvenile and senescent progeroid human mesenchymal progenitor cells (hMPCs) as a cellular aging model.
The researchers discovered large-scale alterations due to global relaxation of structural constraints in the nuclear lamina and the absence of epigenetic markers through in-depth analyses of reorganization and interplay within and between hierarchical layers of the epigenomic organization during hMPC senescence.
They discovered that senescent cells’ epigenome had enhanced epigenetic “entropy” and had poor categorization.
They discovered particular topological conversions that lead to ectopic expression of developmentally limited genes, such as a cluster of pregnancy-specific glycoprotein (PSG) genes, in addition to clarifying the basic relationships between 3D genomic reorganization and transcriptional dysregulation.
Researchers found that separating the PSG cluster from the nuclear lamina and reactivating it can cause cellular senescence in aged hMPCs. PSGs have also been shown to be novel biomarkers of human organismal aging, as well as possible drivers of several forms of cellular and tissue aging.
Overall, the scientists found increasing epigenetic entropy and stimulation of evolutionarily restricted genes to be new characteristics and driving forces of human cellular aging. These discoveries shed new light on the epigenomic foundation of aging regulation. It will also help direct the development of clinical therapies to reduce aging and treat aging-related diseases.
Source:
Journal reference:
Liu, Z., et al. (2022) Large-scale chromatin reorganization reactivates placenta-specific genes that drive cellular aging. Developmental Cell. doi.org/10.1016/j.devcel.2022.05.004.