Reviewed by Danielle Ellis, B.Sc.Jun 6 2022
The formation of aqueous droplets in macromolecules by liquid-liquid phase separation (or coacervation) is a popular topic in life sciences research. DNA is particularly interesting among the numerous macromolecules that form droplets because it is predictable and programmable, both of which are desirable properties in nanotechnology.
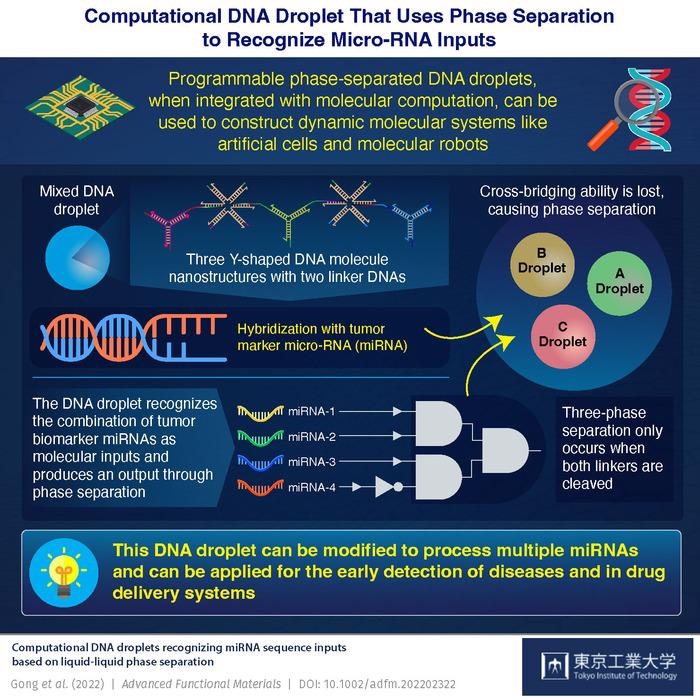
Image Credit: Tokyo Institute of Technology.
The programmability of DNA has recently been used to build and control DNA droplets formed by the coacervation of sequence designed DNAs.
Professor Masahiro Takinoue’s team at Tokyo University of Technology (Tokyo Tech) has created a computational DNA droplet that can identify specific combinations of chemically synthesized microRNAs (miRNAs) that act as tumor biomarkers. Through physical DNA droplet phase separation, the droplets can produce a DNA logic computing output using these miRNAs as a molecular input.
The applications of DNA droplets have been reported in cell-inspired microcompartments. Even though biological systems regulate their functions by combining biosensing with molecular logical computation, no literature is available on integration of DNA droplet with molecular computing.”
Masahiro Takinoue, Study Corresponding Author and Professor, Tokyo University of Technology
The observations were published in the Advanced Functional Materials journal.
A series of experiments were required to create this DNA droplet. To make A, B, and C DNA droplets, researchers first created three types of Y-shaped DNA nanostructures named Y-motifs A, B, and C, each with three sticky ends. Similar droplets tend to stick together naturally, whereas dissimilar droplets require the use of a special “linker” molecule.
To join the A droplet with the B and C droplets, they used linker molecules, which were referred to as AB and AC linkers, respectively.
They tested the “AND” operation in the AB droplet mixture by introducing two input DNAs in the first experiment. The presence of input is recorded as 1 in this operation, while its absence is recorded as 0. The phase separation of the AB droplet mixture took place only at (1,1), that is, when both input DNAs are present, indicating that the AND operation was successfully applied.
Following this research, the investigators decided to add breast cancer tumor markers, miRNA-1 and miRNA-2, to the AC droplet mixture as AND inputs. Because the AND operation was impactful, the computational DNA droplet was able to identify the miRNAs.
The team then illustrated simultaneous AND and NOT operations in an AB mixture with miRNA-3 and miRNA-4 breast cancer biomarkers in subsequent experiments. Finally, they made an ABC droplet mixture and added each of the four breast cancer biomarkers to it. The phase separation in an ABC droplet was determined by the linker cleavage, which resulted in either a two-phase or three-phase separation.
The ability to detect a number of predefined cancer biomarkers or markers for three diseases at the same time was demonstrated by the researchers, thanks to this property of the ABC droplet. Professor Takinoue, who is also the corresponding author, believes that computational DNA droplets have enormous potential.
If a DNA droplet can be developed which can integrate and process multiple inputs and outputs, we can use it in early disease detection as well as drug delivery systems. Our current study also acts as a steppingstone for research in developing intelligent artificial cells and molecular robots.”
Masahiro Takinoue, Study Corresponding Author and Professor, Tokyo University of Technology
Source:
Journal reference:
Gong, J., et al. (2022) Computational DNA Droplets Recognizing miRNA Sequence Inputs Based on Liquid–Liquid Phase Separation. Advanced Functional Materials. doi.org/10.1002/adfm.202202322.