Reviewed by Danielle Ellis, B.Sc.Jun 10 2022
The three types of rotavirus that cause gastroenteritis in individuals, known as groups A, B, and C, are the most well-known and afflict predominantly youngsters.
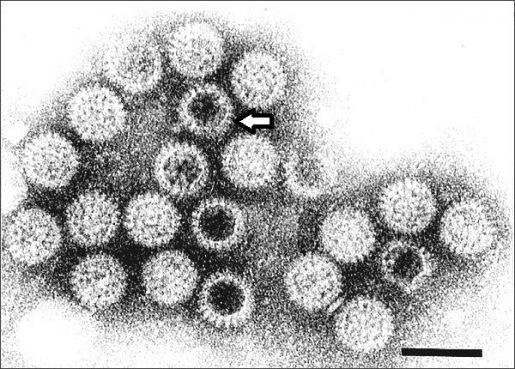 Rotavirus particles visualized by immune electron microscopy. Notice the viral spike indicated by the arrow. Image Credit: Centers for Disease Control
Rotavirus particles visualized by immune electron microscopy. Notice the viral spike indicated by the arrow. Image Credit: Centers for Disease Control
On the other hand, nothing is known about the tip of the virus’s spike protein, called the VP8* domain, which facilitates the infection of gut cells in group B, which primarily affects adults and produces severe diarrhea.
Determining the structure of VP8* in group B rotavirus is important because it will help us understand how the virus infects gastrointestinal cells and design strategies to prevent and treat this infection that causes severe diarrheal outbreaks.”
Dr B. V. Venkataram Prasad, Study Corresponding Author and Professor, Biochemistry and Molecular Biology, Baylor College of Medicine
The team’s initial step was to use X-Ray crystallography to identify the 3D structure of VP8* B, which is a rigorous and time-consuming operation. However, in this circumstance, the typical strategy did not work. The scientists then went to AlphaFold2, a freshly created artificial intelligence-based computing algorithm.
AlphaFold2 predicts the 3D structure of proteins according to their genetic sequence. We knew that the protein sequence of VP8* of rotavirus group B was about 10% similar to the sequences of VP8* of rotavirus A and C, so we expected differences in the 3D structure as well.”
Dr Liya Hu, Study Co-corresponding Author and Assistant Professor, Biochemistry and Molecular Biology, Baylor College of Medicine
Dr Hu added, “But we were surprised when AlphaFold2 predicted a 3D structure for the VP8* B that was not just totally different from that of the VP8* domain in rotavirus A and C, but also that no other protein before had been reported to have this structure.”
With this knowledge, the researchers returned to the lab bench and used X-ray crystallography to validate that the structure of VP8* B predicted by ALphaFold2 corresponded to the actual structure of the protein.
How rotavirus infects cells
Rotavirus A and C infect cells by binding to particular sugar components on histo-blood group antigens, such as the A, B, AB, and O blood groups, which are found in many cells throughout the body, according to a previous study.
It has been suggested that the capacity of different rotaviruses to attach to different sugars on histo-group antigens might explain why some of these viruses infect young children while others infect other people. Unlike VP8* A and VP8* C, the sugar specificity of VP8* B has never been determined before.
Image Credit: Tatiana Shepeleva/Shutterstock.com
Dr Hu stated, “We screened VP8* B against an array of sugars and found that it recognizes N-acetyllactosamine, a sugar common in many cells in the body, that is not recognized by VP8* of rotavirus A and C. Such a 3D structure that also is capable of binding to sugar has not been described before.”
Dr Wilhelm Salmen, co-author of the study and postdoctoral fellow at Baylor College of Medicine adds, “I am excited about identifying a novel 3D protein structure. I am also anticipating all the discoveries that will come from this as we investigate how the new structure interacts with cells to infect them and how this process compares to the one from rotavirus A and C.”
Dr Mary Estes states, “Our lab has been collaborating with Dr. Prasad’s lab for many years to understand the significance of sugar-binding viruses in gastrointestinal infections.”
We can’t cultivate the group B virus yet, but our lab will now try to grow these adult viruses in our human organoid systems, a miniature model of the human gut that can help us probe the mechanism of virus entry and growth. This may lead to new therapeutics that are still needed to treat diarrheal disease.”
Dr Mary Estes, Study Co-author and Cullen Foundation Endowed Chair and Distinguished Service Professor, Virology and Microbiology, Baylor College of Medicine
“This new approach to determine a protein’s 3D structure represents a significant step forward for the field of structural biology,” outlined Dr Hu.
Dr Prasad, who holds the Alvin Romansky Chair in Biochemistry and is a member of the Dan L Duncan Comprehensive Cancer Center concluded, “I am excited about our findings of a new 3D protein structure from an evolutionary point of view. It exemplifies how viruses can evolve by incorporating structurally distinct modules with similar functionality, but how this structure came about in this group B rotavirus is quite intriguing.”
Source:
Journal reference:
Hu, L., et al (2022) Novel fold of rotavirus glycan-binding domain predicted by AlphaFold2 and determined by X-ray crystallography. Communications Biology. doi:10.1038/s42003-022-03357-1