Researchers have developed a novel technique to analyze the composition and quantity of “transfer RNAs” (tRNAs), which are tiny, chemically modified RNAs involved in protein synthesis. Misfolding of tRNAs has been associated with a variety of human diseases, including type 2 diabetes, cancer, and neurological problems.
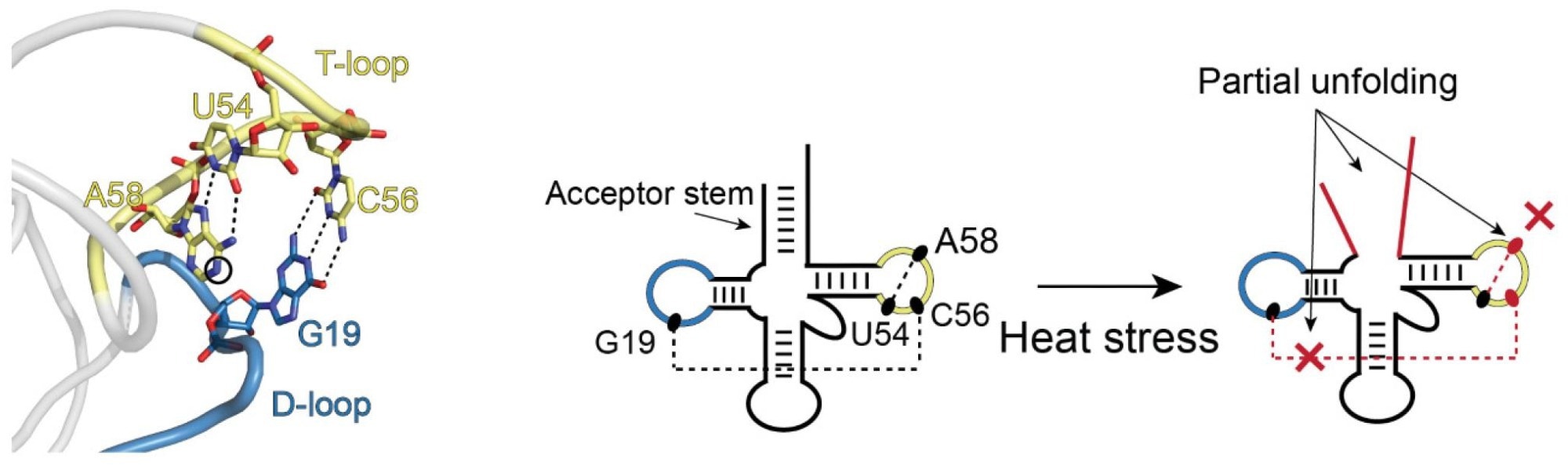 An example of partial unfolding of tRNA under heat stress. The image on the left shows the natural three-dimensional conformation of a portion of the tRNA with the D-loop (blue) and T-loop (yellow) connected in what is referred to as “kissing loops.” The two images on the right show the transition in structure that occurs upon heat stress. D- and T-loops retain their blue and yellow coloring. There is partial unfolding of the acceptor stem and the kissing loops no longer kiss. Image Credit: Bevilacqua Lab, Penn State.
An example of partial unfolding of tRNA under heat stress. The image on the left shows the natural three-dimensional conformation of a portion of the tRNA with the D-loop (blue) and T-loop (yellow) connected in what is referred to as “kissing loops.” The two images on the right show the transition in structure that occurs upon heat stress. D- and T-loops retain their blue and yellow coloring. There is partial unfolding of the acceptor stem and the kissing loops no longer kiss. Image Credit: Bevilacqua Lab, Penn State.
The new technique, which also demonstrates how high temperatures, a common stressor experienced by plants, bacteria, and even humans, can alter tRNA structure, may help in the development of treatments for diseases associated with RNA. It is also applicable to other small, highly modified types of RNA.
An article explaining the approach, created by Penn State academics, was published in the Proceedings of the National Academy of Sciences publication.
About eight years ago, our team developed a method called ‘Structure-seq’ that uses chemical modifications and high-throughput sequencing to determine the structure of messenger RNAs in vivo. We’ve expanded that method here to overcome the challenge of working with smaller RNAs that have very stable structures and are naturally highly modified with chemical tags, like tRNA.”
Philip Bevilacqua, Study Leader and Distinguished Professor, Chemistry, Biochemistry and Molecular Biology, Pennsylvania State University
In many aspects, DNA and RNA are identical. Both are long molecules made up of many small building blocks known as “bases” or “nucleotides,” which in DNA stand for A, T, C, and G (the T is replaced by U in RNA). However, unlike DNA, which consists of two long molecules that run parallel to one another and are joined by hydrogen bonds between complementary units, RNA has only one strand.
This distinction is crucial because RNA molecules fold back on themselves to create brief portions of double-stranded DNA-like bonds that are intermingled with single-stranded segments that create loops and bulges.
It was discovered that tRNA has a basic structure that resembles a cloverleaf with three loops joined by double-stranded segments, all revolving around a central hub. This structure was established in the 1960s and 1970s. Numerous tRNA species exist, and their sequences only differ minimally amongst them.
The structures of the various types of tRNA, which carry particular amino acids—the building blocks of proteins—to the cell’s site of protein synthesis, are essential to their ability to carry out this role.
Ryota Yamagami, who was a postdoctoral researcher at Penn State at the time of the study and is now an assistant professor at Ehime University in Japan noted, “We can predict the structure of RNA molecules based on their sequence and by modifying the molecules with the chemical dimethyl sulfate (DMS), which adds a different chemical tag to certain nucleotides that are exposed in the structure.”
Yamagami also added, “Because tRNAs are small and already highly modified, which can cause problems for sequencing, we had to develop methods to ensure that we captured the full-length molecules.”
According to Yamagami, their process involves treating live cells with DMS before RNA is collected and then chosen by size to separate the tRNA. The RNA molecules must be copied into DNA in order to sequence them. To do this, short DNA segments known as primers are first connected to one end of the RNA molecules. This protects the short RNA sequence’s information from being lost.
When utilized under specific circumstances, a particular form of the enzyme called “reverse transcriptase” does not get inhibited by chemical alterations to the nucleotides, but rather frequently causes mistakes when it comes to a changed nucleotide. The procedure of “mutational profiling” is used by the researchers to determine which nucleotides have been altered in order to aid in their structural predictions.
With this method we get highly accurate structure predictions at the level of individual molecules. This also allows us to determine the abundance of each type of tRNA in the cell and the presence of some natural modifications, which can be important for the efficient production of proteins and the overall health of the cell.”
Philip Bevilacqua, Study Leader and Distinguished Professor, Chemistry, Biochemistry and Molecular Biology, Pennsylvania State University
Through research at the Penn State Huck Institutes of the Life Sciences Metabolomics Core Facility, Jacob Sieg, a chemistry graduate student in the Bevilacqua lab, determined the identification and prevalence of natural changes.
The investigations were carried out on bacterial cells that had been cultivated at normal temperatures, at high temperatures (heat stress), and in cells that had received a brief burst of high temperatures (heat shock). Multiple tRNA molecule types misfolded under circumstances of heat stress and shock, and the relative quantity and structure of various tRNAs varied.
The concept that even a highly structured RNA such as tRNA is conformationally responsive to environmental conditions is an exciting finding from this study. It expands our basic understanding of how cells respond to their environment and stress and this knowledge could help inform the development of novel interventions.”
Sarah M. Assmann, Study Author and Waller Professor, Biology, Pennsylvania State University
The brand-new approach, called “tRNA structure-seq,” is not restricted to tRNA and might be used to investigate the structure of small RNAs linked to human disorders.
“Seeing how tRNA molecules respond to these stresses and beyond will help us better understand how cells respond to stress and could also help us understand diseases linked to tRNA misfolding. This new method overcomes the difficulties of working with small, highly modified RNA molecules and can be applied to any organism and to other small RNAs.” Bevilacqua concluded.
Source:
Journal reference:
Yamagami, R., et al. (2022) Genome-wide analysis of the in vivo tRNA structurome reveals RNA structural and modification dynamics under heat stress. Proceedings of the National Academy of Sciences. doi.org/10.1073/pnas.2201237119.