When the coronavirus that causes COVID-19 infects human cells, the cell’s protein-processing machinery modifies the spike protein, making it more flexible and mobile, potentially increasing its possibility to contaminate other cells and evade antibodies, according to new research from the University of Illinois Urbana-Champaign.
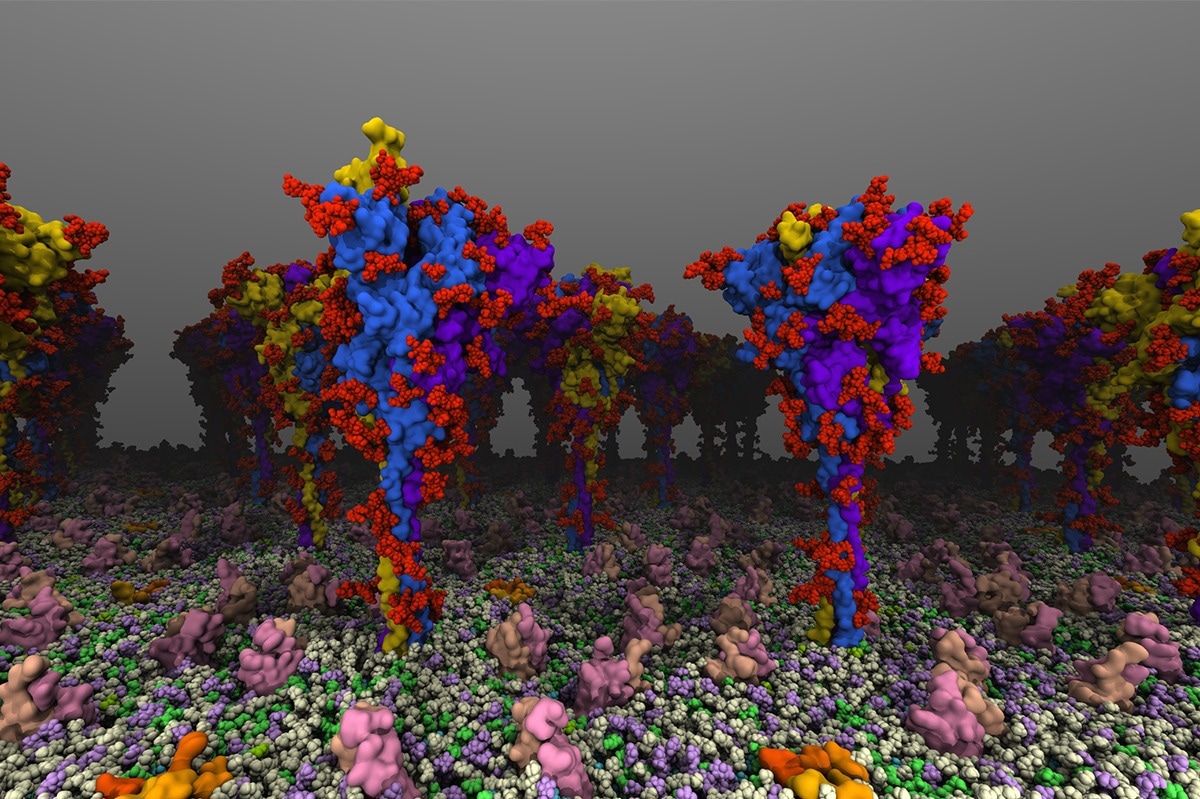 University of Illinois researchers created atomic-level models of the spike protein that plays a key role in COVID-19 infection and immunity, revealing how the protein bends and moves as it seeks to engage receptors. Image Credit: Tianle Chen
University of Illinois researchers created atomic-level models of the spike protein that plays a key role in COVID-19 infection and immunity, revealing how the protein bends and moves as it seeks to engage receptors. Image Credit: Tianle Chen
The researchers developed an atomic-level computational model of the spike protein and drove numerous simulations to investigate the protein’s dynamics and how cell modifications impacted those dynamics. According to the scientists, this is the first investigation to display such a comprehensive picture of the protein that plays a major role in COVID-19 infection and immunity.
Emad Tajkhorshid, a biochemistry professor, postdoctoral researcher Karan Kapoor, and graduate student Tianle Chen released their findings in the journal PNAS.
“The dynamics of a spike are very important—how much it moves and how flexible it is to search for and bind to receptors on the host cell,” said Tajkhorshid, who is also a member of the Beckman Institute for Advanced Science and Technology.
In order to have a realistic representation, you have to look at the protein at the atomic level. We hope that the results of our simulations can be used for developing new treatments. Instead of using one static structure of the protein to search for drug-binding pockets, we want to reproduce its movements and use all of the relevant shapes it adopts to provide a more complete platform for screening drug candidates instead of just one structure.”
Emad Tajkhorshid, Professor, Biochemistry, University of Illinois Urbana-Champaign
The spike protein of SARS-CoV-2, the virus that triggers COVID-19, is the protein that protrudes from the virus’s surface and adheres to receptors on the surface of human cells, allowing the virus to attack them. It is also the focus of antibodies in people who have been immunized or have retrieved from infection.
Numerous studies have been conducted on the spike protein and its amino acid sequence, but the understanding of its structure has primarily been based on static images, according to Tajkhorshid. The atomistic simulations allow scientists to see how the protein engages with receptors on cells it wants to attack and with antibodies that want to attach to it.
They discovered that the protein has many “hinges” or moving parts that enable the protein’s head to twist on the stalk that protrudes from the virus. The researchers documented numerous various conformations of the protein, such as active and inactive forms, and traced how the protein switches between them.
The conformations seen in their computational simulations corresponded to the kinds and frequencies of angles noted in experimental structural characterization, according to the scientists, positively contributing to the simulations’ validity.
The researchers also discovered that host cell processing altered the viral protein’s dynamics. Much attention has been paid to the virus’s genetic code and the mutations that have occurred as new variants have emerged.
However, as it is tucked and “packaged” for transport throughout the cell, the spike protein undergoes a series of changes. The addition of sugars known as glycans at specific points is one of the most prevalent modifications.
Little is known about these post-translational modifications. The main role that has been noted is that glycans shield the protein from the targeting of antibodies. We compared glycosylated and nonglycosylated forms of the spike protein and found significant dynamic differences between the two.”
Tianle Chen, Graduate Student, University of Illinois Urbana-Champaign
The spike protein had a greater range of motion, allowing it to flex and interact with cell surface receptors, according to the scientists. The glycans also conversed with the cell membrane, enabling the spike protein to relocate and look for the receptor along the membrane.
“Glycosylations not only provide an immune shield but also mediate and enhance the mobility of the spikes, increasing the chances of the virus to successfully attach to and infect the human cells. Thus, the functions of these post-translational modifications are much wider than what was initially thought,” Kapoor said. “This understanding can now provide additional opportunities for targeting the function of this virus.”
According to the scientists, their findings suggest the importance of acknowledging not only genetic mutations in the spike protein of new virus variants but also adjustments like glycosylation and how those modifications can contribute to virus infection rates and immune denial. Researchers also predict that other scientists will use their models to create better diagnostics, vaccines, and antiviral drugs.
“The hope is that down the road, this new understanding of the spike protein is going to be useful for therapeutic efforts. I imagine we can target the dynamics of the spike protein with compounds that bind to the hinges and make them inflexible, and therefore in principle, make the virus less effective,” Tajkhorshid said.
Source:
Journal reference:
Kapoor, K., et al. (2022) Posttranslational modifications optimize the ability of SARS-CoV-2 spike for effective interaction with host cell receptors. Proceedings of National Academy of Sciences. doi.org/10.1073/pnas.2119761119.