Scientists from Peking University and the Institute of Genetics and Developmental Biology of the Chinese Academy of Sciences, under the direction of Dr Yuling Jiao, have discovered that gene editing of the transcription factor DUO1 can massively improve wheat grain yield. DUO1 is an APETALA2/ethylene responsive factor (AP2/ERF) transcription factor. The findings were released in Nature Plants.
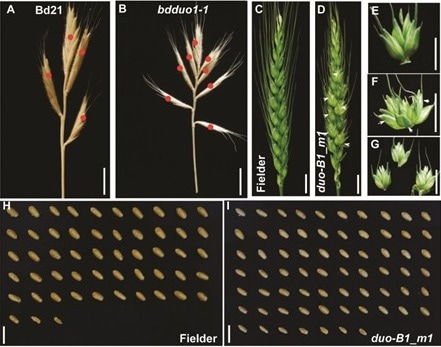 Functional analysis of DUO1. A-B. Brachypodium distachyon wild type (A) with four spikelets on the main spike; mutant (B) with eight spikelets; C-G show the role of this gene in wheat; C. wheat variety Fielder spike type; D. duo1 mutant spike type; E. wheat variety Fielder bearing one spikelet per rachis node; F. mutant bearing three spikelets per rachis node; G. Anatomy of the spike of F; H-I. Comparison of the number of grains in the main spike between wild type Fielder and muant, and the number of grains in the mutant spike is significantly higher. Image Credit: IGDB
Functional analysis of DUO1. A-B. Brachypodium distachyon wild type (A) with four spikelets on the main spike; mutant (B) with eight spikelets; C-G show the role of this gene in wheat; C. wheat variety Fielder spike type; D. duo1 mutant spike type; E. wheat variety Fielder bearing one spikelet per rachis node; F. mutant bearing three spikelets per rachis node; G. Anatomy of the spike of F; H-I. Comparison of the number of grains in the main spike between wild type Fielder and muant, and the number of grains in the mutant spike is significantly higher. Image Credit: IGDB
The discovery of this gene provides superior allelic variation for improving wheat yields and has positive implications for addressing the food security crisis.”
Scott Boden, Professor, Wheat Researcher, University of Adelaide
For tens of thousands of years, humans have modified and domesticated wild plants and animals for their own needs and advancement. One of the most significant dietary staples is wheat, which is an inbred crop with particular evolutionary restrictions and little genetic variation.
The issue of feeding an expanding population is enormous. To further enhance grain yield features in wheat, new alleles must be created and new loci must be found.
A spikelet is located at the base of each of the several axial nodes that make up the compound spike of wheat. Grain number and spikelet number are strongly and favorably associated.
In order to increase wheat grain output, it is therefore both theoretically and practically important to analyse the major genes regulating spikelet number.
In this research, the genes controlling spikelet number in wheat were examined using Brachypodium distachyon spikelet mutant lines.
They discovered the bdduo1 Brachypodium distachyon T-DNA insertion mutant, which has more spikelets per spike. Through a series of genetic studies, they were able to better understand the function of BdDUO1 in the control of spike form in Brachypodium distachyon.
The wheat gene was then modified intentionally by the researchers using CRISPR/Cas9 technology. They discovered that in gene-edited mutants, the spikelets in the lower centre section of the spikes had a multi-spikelet phenotype (two to three spikelets at the base of each rachis node).
In compared to the wild type, live imaging showed more and larger cells in the basal spikelet primordia of the mutant wheat, indicating that the gene controls cell division.
The scientists also learned that the gene-edited mutant wheat had considerably more grains per spike than the wild variety during yield-plot field studies, improving yield per unit area.
Source:
Journal reference:
Wang, Y., et al. (2022) Improving bread wheat yield through modulating an unselected AP2/ERF gene. Nature Plants. doi.org/10.1038/s41477-022-01197-9.