Rady Children’s Institute for Genomic Medicine (RCIGM®) recently announced the publication in Nature Communications of research that describes and examines the efficiency of Genome-to-Treatment (GTRxTM), a computer-controlled, virtual disease management system that combines a quick Whole Genome Sequencing (rWGS®) diagnosis accomplished in 13.5 hours with a custom lab management information system and analysis pipeline.
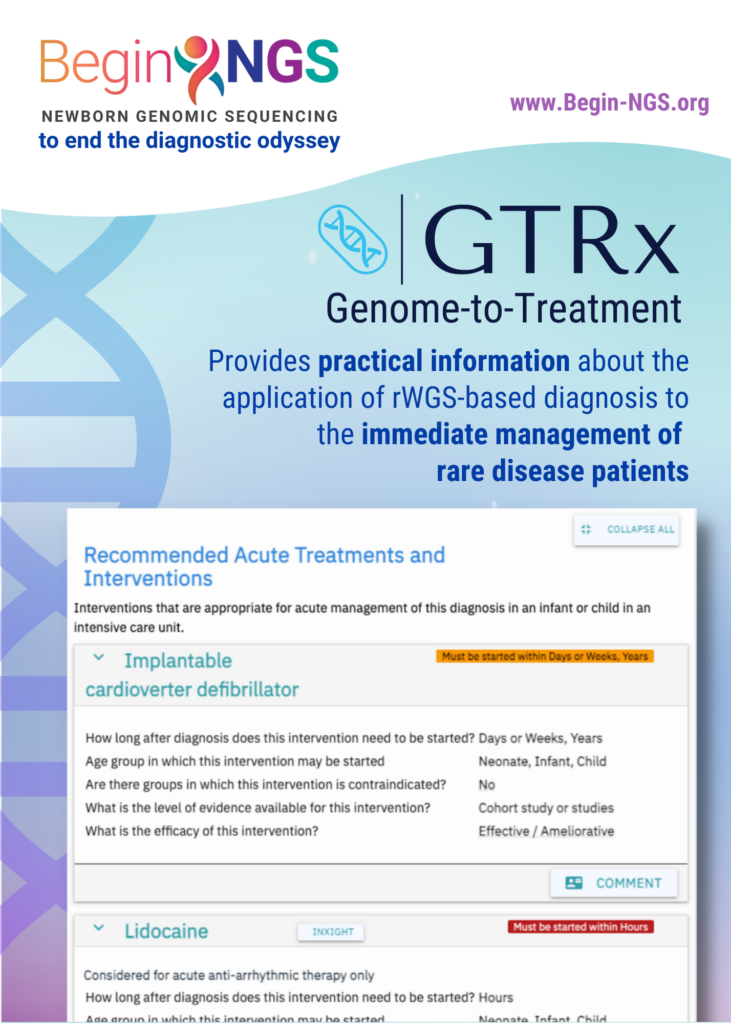
Image Credit: Rady Children’s Institute for Genomic Medicine
GTRx is currently a prototype that contains approximately 500 genetic diseases that progress rapidly and have efficient, treatment options.
Alexion, AstraZeneca’s Rare Disease group, Genomenon, Fabric Genomics, Illumina, SL Data Strategies, Keck Graduate Institute, Rady Children’s Hospital-San Diego, and the UC San Diego departments of Pediatrics and Neuroscience collaborated on the study.
GTRx is intended to provide immediate guidance to frontline neonatologists and intensivists to select optimal therapies for critically ill infants and children in intensive care units as time is of the essence. This study continues to validate the clinical utility of rWGS and how Rapid Precision Medicine™ can improve outcomes for those diagnosed with a rare disease.”
Stephen Kingsmore, MD, DSc, Study Principal Investigator and President, Rady Children’s Institute for Genomic Medicine
Kingsmore was also the CEO of RCIGM.
Nowadays, it takes five years on average and involves several doctors to diagnose a child with a rare disease.
The authors used this information to create a system for computerized genetic illness detection and acute care advice. They started by bringing in numerous advances that improved scalability and reproducibility, including the automated interpretation and enhancing the ability to diagnose genetic illnesses linked with most main classes of genomic variations. This reduced the turnaround time of rWGS to 13.5 hours.
The scientists also used GTRx to give doctors rapid treatment recommendations so they could better comprehend the genetic disorders they had discovered and their potential treatments, which may include medicines, dietary modifications, surgery, medical devices, or other procedures.
GTRx is a key component of the recently unveiled diagnostic and precision medicine guiding tool called BeginNGSTM (roughly “beginnings”), which will use rapid whole genome sequencing (rWGS®) to screen infants for about 400 genetic illnesses for which there are recognized treatments.
In contrast to current pediatric uses of rWGS, which primarily concentrate on children who are already critically ill, BeginNGS uses rWGS to detect and classify treatment options for genetic problems before symptoms appear. It was developed through joint research with Alexion, Fabric Genomics, Genomenon, Illumina, Inc., and TileDB.
BeginNGS employs GTRx to help with clinical decision support after a diagnosis of a rare genetic illness and to lessen variability or delayed adoption of optimal precision treatment.
One of the primary partners in GTRx, Alexion, linked several internet sites for disease data and supplied tools for machine learning to explore the published literature to identify cures for rare genetic diseases.
By integrating tools to enable rapid diagnosis and deliver disease management guidance into an automated virtual system, GTRx is a remarkable example of how we can leverage digital technology to help doctors and patients get answers faster.”
Tom Defay, Alexion’s Deputy Head, Diagnostics Strategy and Development, Rady Children’s Institute for Genomic Medicine
“As a leader in rare diseases for 30 years, Alexion remains committed to helping shorten the diagnostic odyssey for patients, and we are proud of our collaboration to provide RCIGM with technical expertise and support for the development of this innovative technology,” he added.
Illumina, a key collaborator in the creation of GTRx, provided technologies for genome sequencing library preparation, sequencing, analysis, and data transformation.
Ashley Van Zeeland, head of Illumina Open Innovation, says, “We at Illumina are very proud to play a role in this groundbreaking work. Our Customer Collaboration & Innovation team collaborated deeply with Dr Kingsmore and the team to develop solutions that set a new benchmark for enabling end-to-end precision care more quickly for these critically-ill babies.”
An expert clinical panel conducted a comprehensive evaluation of the published data regarding the effectiveness of 9911 currently available medications, technologies, and dietary and surgical therapies for more than 500 serious children’s genetic illnesses as part of the research. The clinical panel concluded that 1527 therapies had sufficient proof of their effectiveness for 421 genetic disorders.
Almost 13 genetic illness information databases, including GARD, Orphanet, and OMIM, were merged with these therapies, efficacy assessments, grades of supporting evidence, and indications and contraindications, and they were attached to diagnostic reports.
“We developed GTRx to both increases the number of children who receive optimal, immediate treatment and to facilitate broader use of rWGS by physicians working in local hospitals who may not be as familiar with genetic diseases as sub- and super-specialists in regional, academic, tertiary or quaternary centers. Our goal is to upskill these physicians by providing patient diagnosis- and disease-specific information in real time,” Dr Kingsmore added.
“Currently GTRx is available for research purposes. Upon validation of clinical utility, we will expand the system to additional genetic diseases and clinical use, as well as incorporating ongoing, expert, open community-based review,” Dr Kingsmore said.
Source:
Journal reference:
Owen, M. J., et al. (2022) An automated 13.5 hour system for scalable diagnosis and acute management guidance for genetic diseases. Nature Communications. doi.org/10.1038/s41467-022-31446-6.