Years of work by Trinity College Dublin developmental biologists in collaboration with colleagues at the University of Edinburgh’s MRC Human Genetics Unit has resulted in an unparalleled resource that will help researchers assess how key genes control the differentiation of tissues and organs in developing embryos.
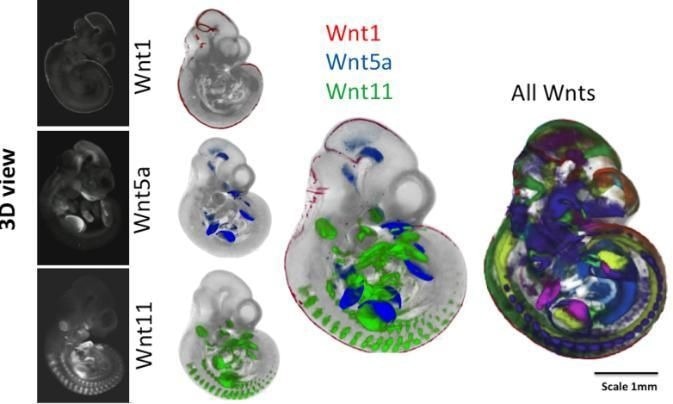 External views of mouse embryos after 10.5 days of embryonic development. The embryos on the left show a technique for revealing where individual genes are expressed (Wnt1, Wnt5a and Wnt11). The middle panel shows the mapping of three gene expression patterns as indicated. On the right, the patterns of all 19 Wnt encoding genes are shown, each in a different color. The embryo space is mapped out by the different expression patterns of these genes, each vital for normal development; a mutation in any one gene can be lethal. Image Credit: Trinity College Dublin.
External views of mouse embryos after 10.5 days of embryonic development. The embryos on the left show a technique for revealing where individual genes are expressed (Wnt1, Wnt5a and Wnt11). The middle panel shows the mapping of three gene expression patterns as indicated. On the right, the patterns of all 19 Wnt encoding genes are shown, each in a different color. The embryo space is mapped out by the different expression patterns of these genes, each vital for normal development; a mutation in any one gene can be lethal. Image Credit: Trinity College Dublin.
The joint team examined these essential genes, their communication links, and their effects in the 3D space of the mouse embryo at various stages of development.
Professor Paula Murphy of Trinity’s School of Natural Sciences elucidated the study and its consequences.
What leap forward has this work taken?
Certain genes and gene networks play an important role in determining how an embryo develops, how different cell types (muscle, bone, neuron) form, and where organs appear. For a few decades now, using molecular biology techniques, we could discover where and when one of these ‘Wnt genes’ is switched on, helping us to understand how it operates in guiding embryonic development.”
Paula Murphy, Professor, School of Natural Sciences, Trinity College Dublin
Paula Murphy adds, “The technique would have to be performed for each gene individually to get a ‘snapshot’ of where it is expressed in 3D embryo space at a particular time. What we have managed to achieve here is the difficult task of mapping these spatial data snapshots for many genes, integrating and comparing where each gene is switched on within the developing embryo.”
“Developing the tools to be able to do this was a major achievement for our computational biology colleagues at the Edinburgh Mouse Atlas project,” stated Paula Murphy.
Were there any discoveries that were surprising?
This approach allowed us to make a number of discoveries that would not have been possible without integrating (mapping) all of the patterns. For example, we could reveal where the pathway is silent because one or other of the components is not activated.”
Paula Murphy, Professor, School of Natural Sciences, Trinity College Dublin
Paula Murphy explains, “Probably the most novel finding was that there are ‘hot spots’ in the embryo where very many of these genes are turned on together. Some of these hot spots are known to be the location of important groups of cells that guide development, but some were previously unknown and reveal previously invisible signaling centers in the embryo.”
What other research question(s) can developmental biologists now ask?
Accompanying this publication, we have made all of the raw and mapped data freely available, as well as the tools for visualizing them so that any developmental biologist can ask questions about this cell communication system in any part of the embryo. For example, colleagues interested in kidney development or brain development can delve in detail into the system components switched on in that part of the embryo.”
Paula Murphy, Professor, School of Natural Sciences, Trinity College Dublin
“We and others can now also use these data to ask questions about how the system has evolved. For example, there are 19 very similar genes in a mouse and a human. These multiple genes have arisen by gene duplication a very long time ago. We can now examine how the genes have diverged, each taking on unique functions during development of a complex multicellular embryo,” added Paula Murphy.
What are the next steps for the team?
Paula Murphy elaborates, “We are currently carrying out focused analysis of the integrated data set in the developing limb buds to understand better how the cell communication system operates in the limb. We also look forward to seeing how the data will be used by colleagues with different specialist interests to further our understanding of all developing systems.”
Source:
Journal reference:
Murphy, P., et al. (2022) Integrated analysis of Wnt signalling system component gene expression. Development. doi.org/10.1242/dev.200312.