Reviewed by Danielle Ellis, B.Sc.Sep 16 2022
The amount of biological data that is currently available has increased exponentially in recent decades as a result of the quick development of the various disciplines in the fields of biological and biomedical research (like genomics, proteomics, and transcriptomics). As an illustration, in just six years, the European Bioinformatics Institute (EMBL-EBI) went from managing a volume of 40 petabytes to dealing with 250 petabytes.
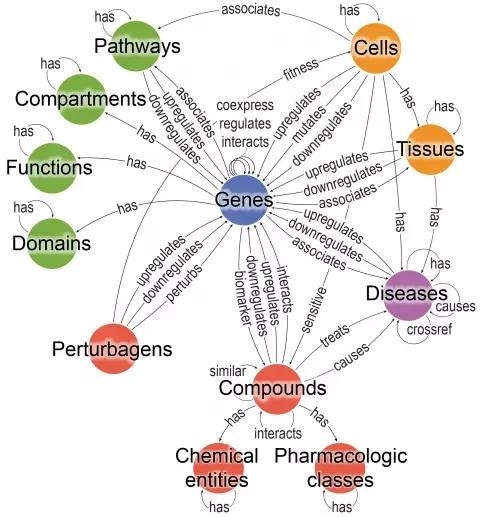 Bioteque is a resource of descriptors for different biological entities. By traversing this knowledge graph through specific entities and relationships we explored more than 1000 paths (aka metapaths) which were encoded into numerical vectors and made available for the community. Image Credit: IRB Barcelona.
Bioteque is a resource of descriptors for different biological entities. By traversing this knowledge graph through specific entities and relationships we explored more than 1000 paths (aka metapaths) which were encoded into numerical vectors and made available for the community. Image Credit: IRB Barcelona.
A computational tool has been created to harmonize, integrate, and simplify this data by researchers working under the direction of Dr Patrick Aloy, an ICREA researcher and head of the Structural Bioinformatics and Network Biology laboratory at IRB Barcelona. The outcome is a knowledge network that details the relationships between various biological entities, including more than 30 million functional interactions.

Image Credit: foxaon1987/Shutterstock.com
The Bioteque functions by combining many layers of biological complexity, allowing it to report on things like two related genes and whether or not they physically interact, are active in the same kind of cells, or are linked to the same disease. Additionally, it can forecast a cell type’s receptivity or resistance to a certain medication.
This computational resource that we've developed is one of the first aimed at unifying biological information and it's the only one to address such diversity and amount of data. It allows access, in an easy and harmonized way, to practically all the biological knowledge currently available, and it has enormous potential to accelerate biomedical research.”
Dr Patrick Aloy, Head, Structural Bioinformatics and Network Biology Laboratory, IRB Barcelona
Almost 1,000 descriptors for 12 biological entities
The Bioteque’s collection of knowledge is organized into 12 different categories of biological things, such as genes, diseases, tissues, cells, etc. The tool takes into account a number of descriptors or features for each of these things, such as a gene’s pattern of mutations, the physical interactions of the proteins produced as a result, the expression of the gene in various cell types, or its association with various disorders.
Around 1,000 different sorts of descriptors are included in the system for the 12 biological entities.
We have worked with information from 150 different databases, so first we had to integrate them, that is, put them all in the same ‘language.’ And then we converted that knowledge into numerical descriptors that could be interpreted by algorithms, and that way we could computationally exploit these networks and connections.”
Adrià Fernández, Study First Author and Doctoral Student, Structural Bioinformatics and Network Biology Laboratory, IRB Barcelona
New databases will be added to the Bioteque on a regular basis as they become available. The tool, databases, and algorithms are all freely accessible and can all be found at https://bioteque.irbbarcelona.org/.
Source:
Journal reference:
Fernández-Torras, A., et al. (2022) Integrating and formatting biomedical data as pre-calculated knowledge graph embeddings in the Bioteque. Nature Communications. doi.org/10.1038/s41467-022-33026-0.