The Babraham Institute’s Reik lab’s study has improved the understanding of the function of DNA methylation during the early stages of development. Technological breakthroughs in this field, such as the capability to collect parallel data types from a single cell and the availability of cell atlas references and comprehensive data sets, are revolutionizing the understanding of the processes that dictate cell fate.
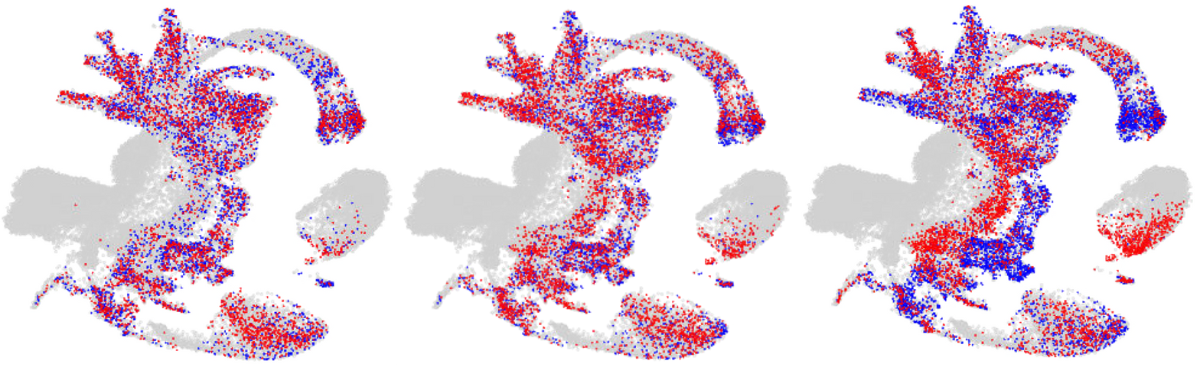
Mapping of cells from the DNMT knock-out mice lines to the reference atlas (grey) with wild-type cells being indicated in blue and knock-out cells being indicated in red. Image Credit: Babraham Institute.
Recognizing the mechanisms that govern the formation of distinct cell types has implications for regenerative medicine as well as understanding developmental disorders and diseases.
The deletion and restoration of DNA methylation are recognized to be critical in establishing cell identity as the embryo’s tissues and organs form. In other circumstances, deleting essential methylation enzymes leads to severe developmental abnormalities and mouse embryo mortality. Despite the significance of DNA methylation in development, the underlying processes by which this happens remain unknown.
This is due to previous constraints on the information scientists could obtain to comprehend the implications of changes in the usual processes of DNA methylation in development, which were limited to the analysis of developmental defects, sample imaging, and limited genome-wide analysis using bulked samples. These approaches were insufficient for resolving effects at the cell type level.
Investigators from the Reik lab at the Institute’s Epigenetics Programme used mice with key methylation enzymes eliminated to do single-cell gene expression studies at the commencement of organ development, which starts at day 8.5 following fertilization.
Using the capabilities of single-cell methodologies, the scientists could track which cell types were impaired in terms of their ability to develop in the mouse embryo, implying the mechanisms underlying the impacts seen at a whole-organism scale.
The study utilized genetically engineered mice in which two significant groups of methylation enzymes were removed. Knock-out mice lines in which individual DNA methyltransferases (DNMT 1, 3a, and 3b) that introduce and maintain DNA methylation were removed, and a system to examine the consequences of a combined deletion of all three TET enzymes (ten-eleven translocation (TET) methylcytosine dioxygenases) 1/2/3 that cause demethylation.
The use of single-cell approaches is really providing the resolution we need to study the mechanics of DNA methylation during development.”
Dr Stephen Clark, Epigenetics Programme, Babraham Institute
Dr Stephen Clark adds, “The picture we were able to build up confirms the repressive role of DNA methylation at this developmental timepoint, firstly that maintaining correct DNA methylation is required to suppress past and alternative cell type identities, and secondly that DNA methylation needs to be removed from parts of the genome to allow certain cell types to form.”
Image Credit: Anusorn Nakdee/Shutterstock
Each mouse line’s gene expression was measured using a technique known as single-cell RNA sequencing. By comparing these expression profiles to a reference dataset, all of the embryo’s cell types were identified.
Following that, the influence of methylation perturbations on cell fate may be determined by comparing the cell type proportions of knock-out embryos (where methylation enzymes were removed) to wild-type embryos at the same stage of development.
The scientists could correlate observed symptoms with impacts on cell type creation at day 8.5 of development and analyze cell-type-specific variations in gene expression that could be related to abnormalities in cell fate commitment.
The ability to both have the whole-organism perspective and the granularity of observing changes in cell types and gene expression has provided us with the ability to tease apart the role of DNA methylation and demethylation in the developing embryo at this particular timepoint to create new insights. It will be equally interesting to apply this approach to later timepoints to understand more about the role of DNA methylation as development progresses.”
Dr Ricard Argelaguet, Study Co-First Author and Former Postdoctoral Researcher, Babraham Institute
The study has developed an interactive data platform offering gene expression readouts at the single-cell level from Dnmt and Tet mutant mice embryos.
This research provides a rich resource to probe the connection between DNA methylation and the establishment of cell fate. This research benefitted from published data sets and reference atlases and we hope that in turn, our work is of use to other researchers in both the development and epigenetics fields.”
Wolf Reik, Professor and Director, Altos Cambridge Institute of Science
Wolf Reik headed the research and is the leader of the Epigenetics Programme at the Babraham Institute.
Source:
Journal reference:
Clark, S. J., et al. (2022) Single-cell multi-omics profiling links dynamic DNA methylation to cell fate decisions during mouse early organogenesis. Genome Biology. doi.org/10.1186/s13059-022-02762-3.