Reviewed by Danielle Ellis, B.Sc.Jan 6 2023
The proper functioning of numerous proteins generated in cells by ribosomes is essential for life. This diversified variety of proteins, called proteome, is sustained by the ribosomes’ vigorous translation elongation of amino acid sequences.
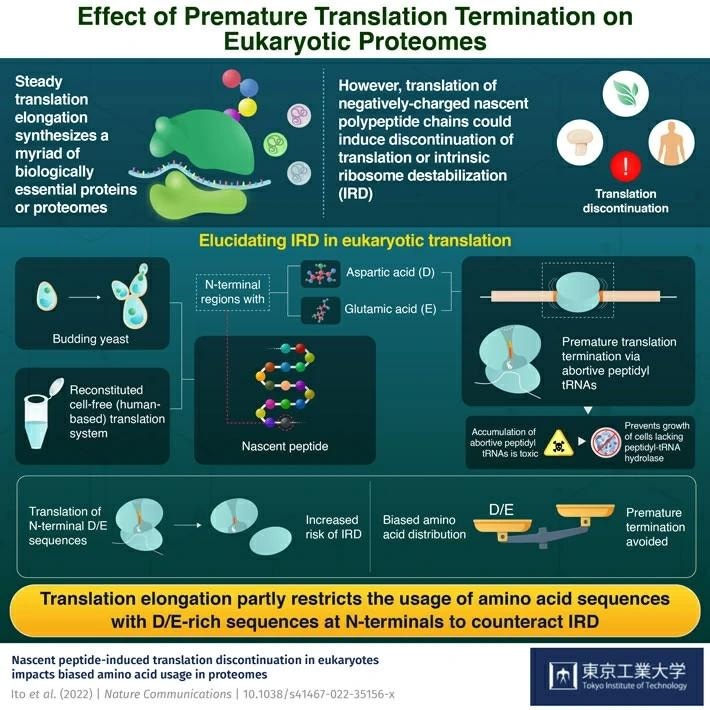
Image Credit: Tokyo Institute of Technology
All living organisms have translation processes that guarantee that nascent chains of polypeptides—long sequences of amino acids—are extended without becoming detached. The rates of elongation, however, are not consistent. Interactions between negatively charged ribosomal RNA and positively charged nascent polypeptides frequently hinder elongation.
According to research, nascent peptide chains not only impede elongation but also destabilize ribosomes in prokaryotic Escherichia coli cells. This sort of premature translation termination is known as intrinsic ribosome destabilization (IRD).
Evidence suggests that nascent peptides with N-terminals rich in aspartic and glutamic acid sequences were mostly responsible for IRD. Because translation systems are conserved, scientists wondered if a similar occurrence could be observed in the cells of eukaryotic organisms, including plants, fungi, and animals.
Image Credit: Meletios Verras/Shutterstock.com
A group of scientists from Japan headed by Prof. Hideki Taguchi of the Tokyo Institute of Technology (Tokyo Tech) was recently successful in proposing some solutions to this topic. The scientists investigated the IRD phenomena in eukaryotes using budding yeast cells and a reconstructed cell-free translation system in their latest paper published in Nature Communications.
Previous studies have explored the impact of aspartic acid and glutamic acid sequences on bacterial ribosomal translation. However, not much is about eukaryotic cells. So, we chose a eukaryotic organism like yeast to investigate the premature termination of translation and if there were any mechanisms present to counter IRD.”
Hideki Taguchi, Study Corresponding Author and Professor, Tokyo Institute of Technology
The researchers revealed that, similar to bacteria, nascent peptide chains with aspartic acid (D) or glutamic acid (E) in their N-terminal regions caused IRD to terminate translation in yeast cells. They also discovered that peptidyl-tRNA buildup reduced cell growth in yeast missing peptidyl-tRNA hydrolase, an essential cellular enzyme.
The peptidyl-tRNAs produced by IRD are cleaved by peptidyl-tRNA hydrolase, which recycles the peptidyl-tRNAs outside the ribosome complex. The accumulation of these abortive peptidyl-tRNAs is toxic, since yeast lacking the enzyme cannot grow when IRD-prone sequences are overexpressed.”
Hideki Taguchi, Study Corresponding Author and Professor, Tokyo Institute of Technology
The group's bioinformatics analysis, on the other hand, found a novel way yeast cells could minimize the risk of IRD. They discovered a biased amino acid distribution in the proteomes, where the translation elongation process preferred amino acid sequences having D/E runs in their N-terminal region.
This research sheds new light on the eukaryotic cell’s elongation dynamics and the mechanisms in place to limit translation errors during protein synthesis.
Understanding the factors that affect overall amino acid usage in proteomes can help us improve the expression of recombinant proteins. This is essential for the production of useful proteins that can have clinical and industrial applications.”
Hideki Taguchi, Study Corresponding Author and Professor, Tokyo Institute of Technology
Source:
Journal reference:
Ito, Y., et al. (2022) Nascent peptide-induced translation discontinuation in eukaryotes impacts biased amino acid usage in proteomes. Nature Communications. doi.org/10.1038/s41467-022-35156-x.