In a new study, Ran Elkon and Ruth Ashery-Padan of Tel Aviv University, Israel, and colleagues discovered a new genetic risk factor for adult-onset macular degeneration (AMD) by combining a map of gene regulatory sites with disease-associated loci.
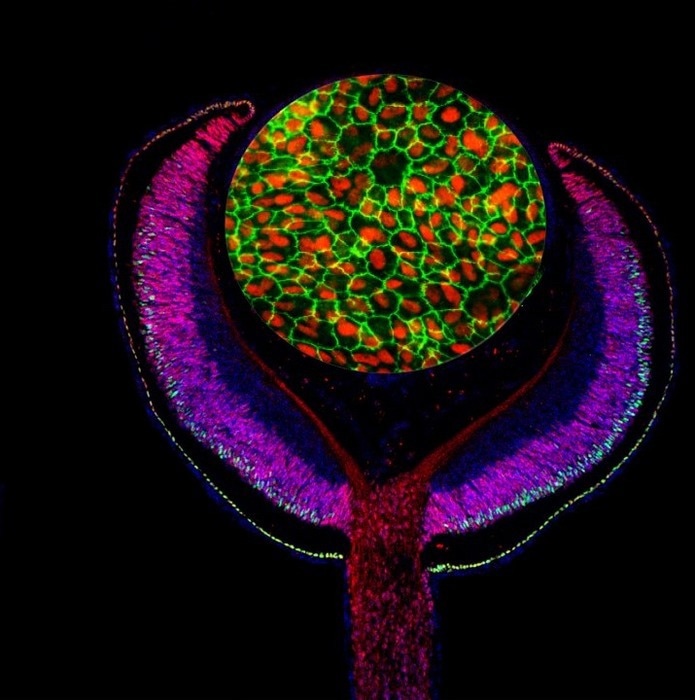 Composite of an embryonic mouse eye cup (E14.5) labeled with antibodies against the developmental transcription factors Lh×2 (red) and Ot×2 (green), and cultured human retinal pigmented epithelium (RPE) labeled with antibodies against MITF (red) and ZO-1 (green). Image Credit: Images by Mazal Cohen-Gulkar, composite by Ruth Ashery-Padan
Composite of an embryonic mouse eye cup (E14.5) labeled with antibodies against the developmental transcription factors Lh×2 (red) and Ot×2 (green), and cultured human retinal pigmented epithelium (RPE) labeled with antibodies against MITF (red) and ZO-1 (green). Image Credit: Images by Mazal Cohen-Gulkar, composite by Ruth Ashery-Padan
The results were published on January 17th, 2023 in the open-access journal PLOS Biology. The discovery broadens the understanding of the main factor causing adult visual impairment.
A layer of tissue sandwiched between the photoreceptors, which receive light, and the choriocapillaris, which feeds the retina, called the retinal pigmented epithelium (RPE), is dysfunctional in AMD.
Given the central role of the RPE in AMD, the authors started by investigating a transcription factor called LH×2, which is crucial for RPE development according to the team’s analysis of mouse mutants.
The majority of the affected genes were down-regulated when LH×2 activity was knocked down in RPE derived from human stem cells, indicating that LH×2’s function was probably that of a transcriptional activator, binding to regulatory sites on the genome to increase the activity of other genes.
The researchers discovered that LH×2 and one impacted gene called OT×2 worked together to control numerous genes in the RPE. The mapping of the genomic sites that OT×2 and LH×2 could bind to has revealed that 68% of those sites (864 sites in total) that bound LH×2 was also bound by OT×2, indicating that they probably cooperate to promote the activity of a large number of genes involved in RPE development and function.
A genome-wide association study (GWAS), which identifies genome sequence variations between individuals (known as single nucleotide polymorphisms, or SNPs) that co-occur with the disease, is a common technique for discovering genes that could contribute to disease.
Such studies in AMD have been conducted before in a large number. A GWAS cannot, however, by itself identify a causal mechanism.
In this case, the authors used their LH×2/OT×2 binding data and GWAS data to focus on variations that affected the transcription factors’ binding and may, therefore, be linked to disease.
The promoter region of a gene called TRPM1, which had previously been linked to AMD, contained one such binding site.
Researchers discovered that the sequence variant at that site changed the strength of LH×2’s binding; the C version bound it more strongly than the T version, and TRPM1 gene activity was higher when the C allele was present instead of the T allele.
The study’s findings suggest that the previously recognized elevated risk of AMD associated with the variant discovered in the GWAS was caused by a decrease in the binding affinity of the LH×2 transcription factor for the TRPM1 gene promoter, which in turn resulted in a decrease in the gene’s activity.
Previous research has demonstrated that the gene’s mutations also result in visual impairment because the gene encodes a membrane ion channel.
Our study exemplifies how delineation of tissue-specific transcriptional regulators, their binding sites across the genome, and their downstream gene-regulatory networks can provide insights into a complex disease’s pathology.”
Ruth Ashery-Padan, Researcher, Tel Aviv University
Source:
Journal reference:
Cohen-Gulkar, M., et al. (2022). The LHX2-OTX2 transcriptional regulatory module controls retinal pigmented epithelium differentiation and underlies genetic risk for age-related macular degeneration. PLoS Biology. doi.org/10.1371/journal.pbio.3001924