Designing genome editing strategies for each patient’s mutation would be impractical and expensive because many genetic diseases are induced by diverse mutations spread across an entire gene.
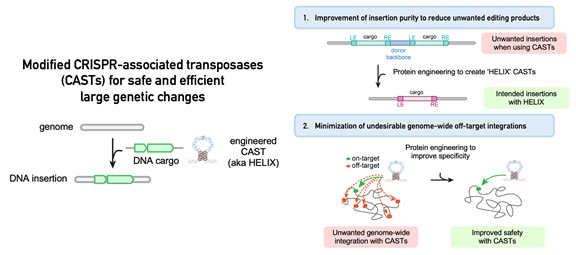
Image Credit: Massachusetts General Hospital
Recently, researchers at Massachusetts General Hospital (MGH) created an improved technique that increases the precision of inserting large DNA segments into a genome.
This method could be used to insert a whole “wild-type” or normal replacement gene, acting as a blanket therapy for a disease regardless of a patient’s specific mutation.
Through the use of a reprogrammable guide RNA, a promising new class of technologies known as CRISPR-associated transposases (CASTs) that can easily target large DNA insertions to desired genomic sites is being improved.
However, in their natural state, CASTs have undesirable characteristics that make them unsuitable for genome editing applications. These characteristics include poor product purity (the frequency with which the intended DNA sequence is inserted into the genome) and a comparatively high rate of undesired off-target integration at unintended sites in the genome.
A group led by senior author Ben Kleinstiver, PhD, an assistant investigator in the Center for Genomic Medicine at MGH and an assistant professor at Harvard Medical School, and first author Connor Tou, a graduate student at MIT and MGH, overcame these drawbacks by using protein engineering techniques to alter the characteristics of CAST systems, which appeared in a study published in Nature Biotechnology.
They discovered that supplementing CASTs with a specific enzyme known as a nicking homing endonuclease significantly improved product purity toward the intended insertion.
The structure of CASTs was further optimized, resulting in DNA insertions at intended genomic targets with high integration efficiency and significantly fewer insertions at undesirable off-target sites.
The name “HELIX,” which stands for Homing Endonuclease-assisted Large-sequence Integrating CAST-compleX, was given to the new and improved system by the researchers.
We demonstrated a generalizable approach that can be used to modify a variety of CAST systems into safer and more effective versions that have high product purity and genome-wide specificity. By combining our insights, we created HELIX systems with greater than 96% on-target integration specificity—increased from approximately 50% for the naturally occurring wild-type CAST system. We also determined that HELIX maintains its advantageous properties in human cells.”
Connor Tou, Study First Author and Graduate Student, Massachusetts Institute of Technology
According to Kleinstiver, the technology could be used for purposes other than reintroducing healthy genes into people who have mutations that cause disease.
Additionally, programmable DNA integration can facilitate cell engineering efforts where installation of large genetic sequences at targeted locations could endow cells with new capabilities while obviating safety, efficacy, and manufacturing issues resulting from traditional random integration approaches.”
Ben Kleinstiver, PhD, Study Senior Author and Assistant Investigator, Center for Genomic Medicine, Massachusetts General Hospital
Source:
Journal reference:
Tou, C. J., et al. (2022). Precise cut-and-paste DNA insertion using engineered type V-K CRISPR-associated transposases. Nature Biotechnology. doi.org/10.1038/s41587-022-01574-x