Methanogens are microbes that produce methane when there is little or no oxygen in their surroundings. Their methane production—for instance, in ruminant digestive tracts—is important for global carbon cycling because methane is a powerful greenhouse gas, but it can also be employed as an energy source to heat homes.
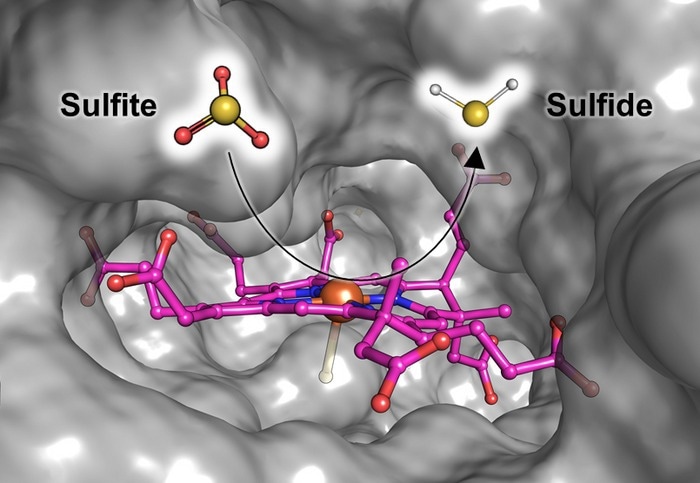 Illustration of Fsr’s catalytic site where sulfite gets reduced to sulfide. The siroheme (in pink) that binds and converts the sulfite is embedded in a cavity of the protein (gray surface) which is solvent accessible. This way, the sulfite can easily enter the protein and the produced sulfide can leave it. Image Credit: Max Planck Institute for Marine Microbiology
Illustration of Fsr’s catalytic site where sulfite gets reduced to sulfide. The siroheme (in pink) that binds and converts the sulfite is embedded in a cavity of the protein (gray surface) which is solvent accessible. This way, the sulfite can easily enter the protein and the produced sulfide can leave it. Image Credit: Max Planck Institute for Marine Microbiology
A toxic base for growth
The study’s objects are two marine heat-loving methanogens: Methanothermococcus thermolithotrophicus (which thrives in geothermally heated sediments at around 65 °C) and Methanocaldococcus jannaschii (which favors deep-sea volcanos at around 85 °C). They acquire cellular energy by producing methane and obtain sulfur for growth in the form of sulfide from their environment.
While sulfide is toxic for many organisms, methanogens require it and can endure high concentrations. Their Achilles’ heel, however, is the poisonous and reactive sulfur compound sulfite, which annihilates the enzyme required to produce methane.
Both researched organisms are occasionally subjected to sulfite in their environments, for instance, when oxygen enters and reacts with the reduced sulfide. Due to the formation of sulfite as a result of its partial oxidation, methanogens must safeguard themselves. But how are they able to do so?
A molecular snapshot of the process
Marion Jespersen and Tristan Wagner from the Max Planck Institute for Marine Microbiology in Bremen, Germany, along with Antonio Pierik from the University of Kaiserslautern, have now provided a glimpse of the enzyme detoxifying the sulfite.
The F420-dependent sulfite reductase, or Fsr, is a butterfly-shaped enzyme. It is capable of converting sulfite to sulfide, a safe source of sulfur required by methanogens for growth. Jespersen and her coworkers describe how the enzyme works in the current study.
The enzyme traps the sulfite and directly reduces it to sulfide, which can be incorporated, for example, into amino acids. As a result, the methanogen doesn’t get poisoned and even uses the product as its sulfur source. They turn poison into food!”
Marion Jespersen, Max Planck Institute for Marine Microbiology
It appears to be straightforward. But Jespersen and her coworkers reported an intriguing and complicated overlap.
There are two ways of sulfite reduction: dissimilatory and assimilatory. The organism under study uses an enzyme that is built like a dissimilatory one, but it uses an assimilatory mechanism. It combines the best of both worlds, one could say, at least for its living conditions.”
Marion Jespersen, Max Planck Institute for Marine Microbiology
It is hypothesized that enzymes from both the dissimilatory and assimilatory pathways evolved from a common ancestor.
Sulfite reductases are ancient enzymes that have a major impact on the global sulfur and carbon cycles. Our enzyme, the Fsr, is probably a snapshot of this ancient primordial enzyme, an exciting look back in evolution.”
Tristan Wagner, Head, Max Planck Research Group Microbial Metabolism, Max Planck Institute for Marine Microbiology
Biotechnological applications in view
The Fsr not only opens up evolutionary possibilities, but it also helps comprehend the fascinating world of marine microorganisms. Methanogens that can only grow on sulfite avoid the risks of sulfide, their usual sulfur substrate.
Wagner states, “This opens opportunities for safer biotechnological applications to study these important microorganisms. An optimal solution would be to find a methanogen that reduces sulfate, which is cheap, abundant, and a completely safe sulfur source.”
This methanogen, Methanothermococcus thermolithotrophicus, already exists. The investigators hypothesized that Fsr instigates the final reaction of this sulfate reduction pathway because sulfite is one of its intermediates.
“Our next challenge is to understand how it can transform sulfate to sulfite, to get a complete picture of the capabilities of these miracle microbes,” Wagner concludes.
Source:
Journal reference:
Jespersen, M., et al. (2023) Structures of the sulfite detoxifying F420-dependent enzyme from Methanococcales. Nature Chemical Biology. doi.org/10.1038/s41589-022-01232-y.