A biological sample contains thousands of unique cells that may all be examined separately and individually, cell by cell. They can be grouped into clusters based on the activity of their genes. But which genes—those referred to as its “marker genes”—are particularly distinctive of a certain cluster? The identification and analysis of these marker genes are facilitated by a novel statistical technique known as the Association Plot.
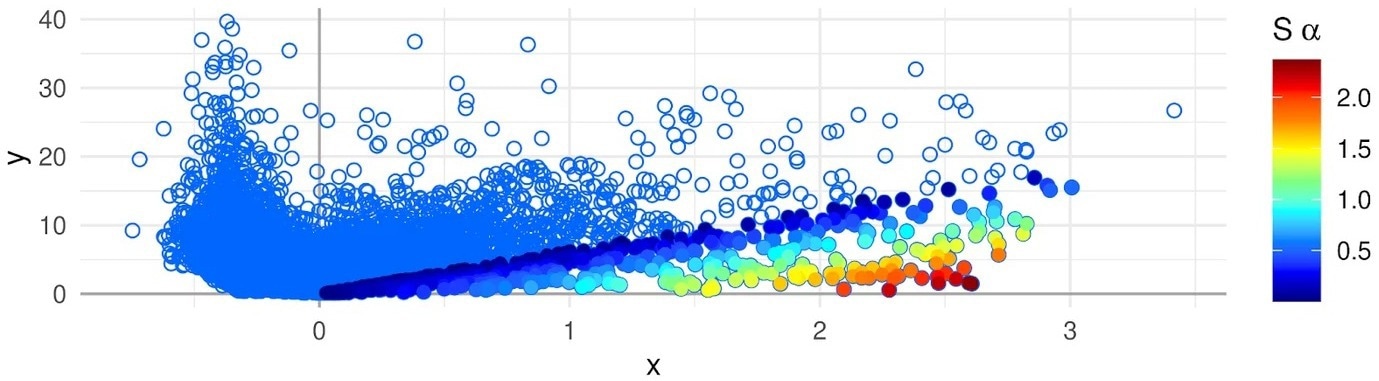 Association Plot of a cell cluster that has been generated with the APL package–genes are represented with dots. The color scale on the right side represents the strength of the gene-cluster association. Genes colored in red are the most cluster-specific. Image Credit: E. Gralinska, Max Planck Institute for Molecular Genetics.
Association Plot of a cell cluster that has been generated with the APL package–genes are represented with dots. The color scale on the right side represents the strength of the gene-cluster association. Genes colored in red are the most cluster-specific. Image Credit: E. Gralinska, Max Planck Institute for Molecular Genetics.
Which genes “mark” the identity of a certain cell type by being particular to that cell type? These days, it might be difficult to provide a response due to the growing amount of databases. Marker genes are frequently the only genes that have been identified in certain cell populations. However, many more genes could be unique to a certain cell type but have not yet been identified.
Finding its flag genes is made simpler by “Association Plots (APL),” a novel statistical technique for displaying gene activity inside a cell cluster. The graphs compare a specific cluster’s gene activity to that of every other cluster in the data set. They also make it simple to discover which genes are shared by different clusters.
Association Plots not only allow us to identify new marker genes. It also works the other way around—we are able to match clusters of unknown identity in a dataset to cell types, based on a provided list of marker genes.”
Elzbieta Gralinska, PhD Student, Department of Computational Molecular Biology, Max Planck Institute for Molecular Genetics
The biotechnologist is a member of Martin Vingron’s team, which created the technique, tested it on two datasets that were made accessible to the public, and then published the findings in the Journal of Molecular Biology. APL has also been made available as a free module for the R statistical environment. Researchers may use the APL software to visually check their single-cell data and choose certain genes with the cursor to get more specific information.
Analyzing and grouping single cells
What makes it essential in the first place to find marker genes? Individual RNA molecules in distinct cells can be decoded using current sequencing technology. A sample of each cell’s RNAs can be extracted from a blood sample, for instance, and decoded. The active genes that were translated into RNA molecules are represented by this single-cell data.
The advantage: It is possible to identify the cell from where a certain RNA originated, eliminating the problem of which cell type it belongs to. The disadvantage: Out of tens of thousands of cells, sequencing thousands of RNAs results in incredible volumes of data.
Sorting the cells according to their RNA concentration is one solution.
Single-cell data are composed of a wild mix of many different cell types. We are interested in cells of the same cell type, which should all behave similarly. For us, the marker genes define a cell type.”
Dr Martin Vingron, Professor, Scientific Member and Director, Department of Computational Molecular Biology, Max Planck Institute for Molecular Genetics
Consequently, it is possible to computationally group comparable cells in a logical manner.
Explore cell clusters interactively
The scientists illustrated how the new algorithm functions using white blood cell data that is openly available. T-cells, B-cells, and monocytes are only a few of the several varieties of white blood cells that are organized into discrete clusters. The researchers were able to demonstrate that close relatives among the blood cells also share a high resemblance in their gene activity and corroborated known flag genes.
Each of the marker genes we found with APL could have been discovered by at least one other existing method for identification of marker genes. Existing tools provide long lists of genes and score values. Oftentimes, users go through the list and stop at an arbitrary cut-off.”
Elzbieta Gralinska, PhD Student, Department of Computational Molecular Biology, Max Planck Institute for Molecular Genetics
However, Gralinska claims that APL has a graphical representation of the outcomes that give it an edge over the current techniques.
In contrast, the new approach offers a means to view these genes, click on each one, and examine its activity more closely, Gralinska adds. “We’re not just providing lists of marker genes, we're allowing users to review how these genes behave,” the researcher notes.
Gralinska says, “With Association Plots, they can dive into their data to learn more about each cell type.” Additionally, Gralinska explains that using the APL software’s Gene Ontology terms enrichment analysis, it is relatively simple to decipher the biological function of the most intriguing genes in a future step. Gralinska views this as “a very useful feature.”
The underlying mathematical model
Visual representation of high-dimensional data containing information on gene activity is not possible without sacrificing information. The same is true with clustered data, which makes analysis more difficult. “Our trick is that we take into account many more than just two or three dimensions, but ultimately create a two-dimensional diagram,” Gralinska states.
The Association Plots are generated from a mathematical method that embeds genes and cells in the same high-dimensional space at the same time. In this space, measuring the distances between genes and a certain cell cluster yields pairs of values that indicate a gene’s affiliation with that cluster and provides information about its association with other clusters.
“One shortcoming of APL is that we rely on pre-clustered data, which means we have to rely on other techniques for clustering. Nevertheless, we hope that our new method will find many new users. We find that a visual and interactive process simply makes a better analysis,” Martin Vingron concludes.
Source:
Journal reference:
Gralinska, E., et al. (2022) Visualizing Cluster-specific Genes from Single-cell Transcriptomics Data Using Association Plots. Journal of Molecular Biology. doi.org/10.1016/j.jmb.2022.167525.