Amyotrophic lateral sclerosis (ALS), sometimes referred to as Stephen Hawking’s disease and Lou Gehrig’s disease is a neurological condition that causes the body’s muscles to gradually lose control. Over 90% of cases have an unknown etiology, however, both genetic and environmental factors are suspected to be involved. It is currently incurable.
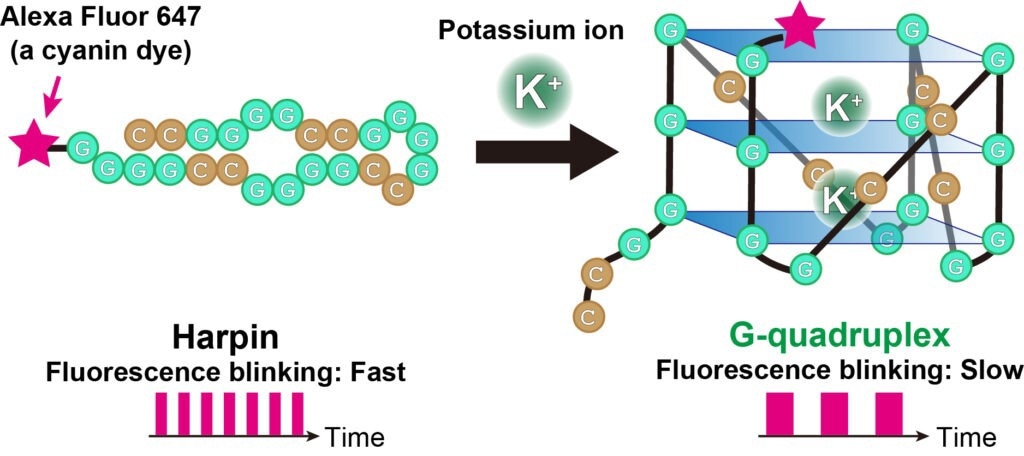 The fluorescent blinking of cyanine dye (Alexa Fluor 647, pink star) bound to RNA changes depending on the structure of the RNA. When the RNA is folded like a hairpin, the fluorescent blinking is fast, and when the RNA switches to a G-quadruplex, the blinking is slow. Image Credit: Akira Kitamura
The fluorescent blinking of cyanine dye (Alexa Fluor 647, pink star) bound to RNA changes depending on the structure of the RNA. When the RNA is folded like a hairpin, the fluorescent blinking is fast, and when the RNA switches to a G-quadruplex, the blinking is slow. Image Credit: Akira Kitamura
The research teams of Prof. Jerker Widengren at the KTH Royal Institute of Technology in Sweden and Dr Akira Kitamura at the Faculty of Advanced Life Science at Hokkaido University have created a revolutionary method that can recognize an RNA’s distinctive structure in real time in living cells.
The fluorescence-microscopic spectroscopy-based method was described in the journal Nucleic Acids Research.
One of the genetic factors that is believed to be involved in the development of ALS is a specific sequence of RNA that forms a four-stranded structure, called a G-quadruplex. Normally, these structures regulate the expression of genes. However, a mutation in chromosome 9 in humans results in the formation of G-quadruplexes that may play a role in neurodegenerative diseases including ALS.”
Dr Akira Kitamura, Senior Lecturer, Faculty of Advanced Life Science, Hokkaido University
The inability to see the production and placement of G-quadruplexes in living cells in real time has been one of the largest obstacles to knowing the precise function of G-quadruplexes in disease. The Kitamura and Widengren groups were successful in creating a straightforward, reliable, and broadly applicable approach that addresses current problems.
The method monitors a cyanine dye known as Alexa Fluor 647 (AF647). When a dye is attached to RNA, the formation of RNA G-quadruplexes changes the dye’s fluorescence blinking state.
To identify this fluorescence blinking in real time, the teams examined the AF647-labeled RNA using a microscopy method known as TRAST (TRAnsient STate Monitoring).
Dr Kitamura added, “Visually, the time-resolved changes in intensity of fluorescence appear as blinking. In TRAST, we expose cells to a specific pattern of changing light intensities and measure the average intensity of fluorescence emitted from the RNA-bound dye in the cells across specific time intervals. By measuring changes in blinking properties, we can distinguish the structures of RNA within the cell.”
Under controlled laboratory conditions, the scientists calibrated their experiment and identified precisely which fluorescent blinking represented RNA G-quadruplexes. This insight allowed them to use TRAST to locate RNA G-quadruplexes within live cells.
The research presented here demonstrates that cyanine dyes can offer sensitive readout parameters on the RNA G-quadruplex folding states in live cells, even for single cells. The ability to analyze RNA G-quadruplexes in disease in real time at the intracellular level is thereby made possible. It could also be used to research how proteins in cells fold and misfold.
Source:
Journal reference:
Kitamura, A., et al. (2023). Trans-cis isomerization kinetics of cyanine dyes reports on the folding states of exogeneous RNA G-quadruplexes in live cells. Nucleic Acids Research. doi.org/10.1093/nar/gkac1255