Using high-resolution cryo-electron microscopy (cryo-EM), UAB researchers determined the structure of amyloid fibers formed by the protein hnRNPDL-2, which has been linked to limb-girdle muscular dystrophy type 3 and concluded that the protein’s inability to form amyloid fibers, rather than aggregation, would be the cause of the disease.
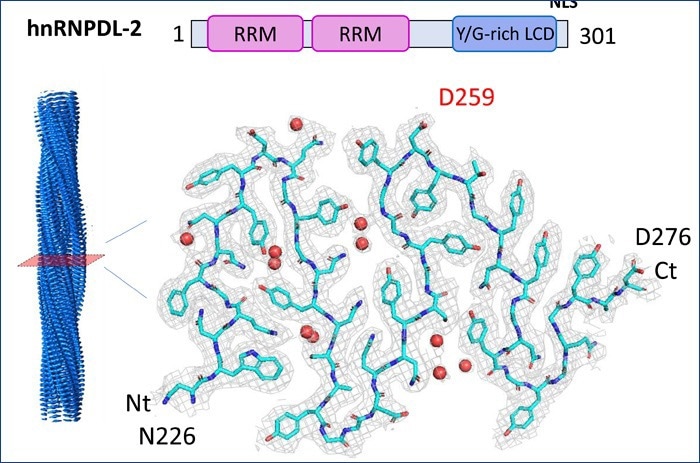 Structure of hnRNPDL-2 amyloid fibers obtained by cryoEM at 2.5 A resolution. The upper part shows the organization of the protein with two nucleic acid binding domains in pink and a low-complexity domain responsible for its assembly. The lower part shows the cryoEM map obtained and the structure of an amyloid fiber layer and its mutations, to better understand its implication in health and disease. Image Credit: Universitat Autònoma de Barcelona
Structure of hnRNPDL-2 amyloid fibers obtained by cryoEM at 2.5 A resolution. The upper part shows the organization of the protein with two nucleic acid binding domains in pink and a low-complexity domain responsible for its assembly. The lower part shows the cryoEM map obtained and the structure of an amyloid fiber layer and its mutations, to better understand its implication in health and disease. Image Credit: Universitat Autònoma de Barcelona
A Spanish research group has determined the first amyloid structure at high resolution. The investigation, published in Nature Communications, directs the treatment toward the quest for molecules that stabilize or enable amyloid formation and opens the door to studying other functional amyloids and their mutations using the same technique in order to better understand their role in health and disease.
Limb-girdle muscular dystrophy type 3 (LGMD D3) is an uncommon condition that is characterized by increasing muscle weakness caused by point mutations in the hnRNPDL-2 protein. It is a little-known protein that belongs to the RNA-associated ribonucleoprotein (RNP) family and has the ability to assemble to form functional amyloid structures.
Amyloids are formed by thousands of pieces of the same protein joining together to form very stable and structured fibers (protein aggregates). Their formation is frequently associated with diseases like Parkinson’s and Alzheimer’s, but they are also used for functional purposes by various organisms, though the number of functional amyloids described in humans remains small.
In a study published in Nature Communications, investigators from the Universitat Autònoma de Barcelona (UAB) ascertained the structure of amyloid fibers formed by the protein hnRNPDL-2.
Their structure and activity indicate that they are stable, non-toxic amyloid fibers that bind nucleic acids when aggregated. The findings suggest that LGMD D3 could be a protein loss-of-function disease, with the pathology caused by the inability to form the amyloid structures described in the study.
Our study challenges the hypothesis that the aggregation of this protein is the cause of the disease and proposes that it is the inability to form a fibrillar structure that has been selected by evolution to bind nucleic acids that causes the pathology.”
Salvador Ventura, Professor and Researcher, Biochemistry and Molecular Biology, Institute of Biotechnology and Biomedicine
Salvador Ventura led the research along with the first author of the paper, Javier Garcia-Pardo, a Juan de la Cierva-Incorporación researcher at IBB-UAB.
Utilizing high-resolution cryo-electron microscopy (cryo-EM), researchers determined the structure of the hnRNPDL-2 protein’s amyloid fibers. This is the first complete structure of human functional amyloid solved using this technique; earlier, only structures created by fragments of these proteins had been solved. It is also the first high-resolution amyloid structure determined by a Spanish research team.
The protein’s structure varies from that of other pathological amyloid proteins in that it encompasses a highly hydrophilic nucleus—the amino acid linked to LGMD D3. However, here, compared to other diseases, amyloid formation is not toxic but is vital for protein function.
According to the investigators, the findings alter the concept of the origin of the disease and the ways in which it should be treated.
Previously, we thought that, as in many neurodegenerative diseases, LGMD D3 originated because mutations in patients caused the initially soluble protein to form aggregates and, therefore, the search for anti-aggregant molecules could be a potential therapy. Now we know that this would be a mistake, since it is the incorrect formation of the fiber that seems to trigger the disease; therefore, molecules that stabilize this structure or facilitate its formation would be the most appropriate.”
Salvador Ventura, Professor and Researcher, Biochemistry and Molecular Biology, Institute of Biotechnology and Biomedicine
Understanding the molecular structures of amyloids
Because some human amyloids can aggregate in both functional and pathological ways, it is critical to understand their molecular structures to ascertain their distinct qualities and functions.
RNPs like the one investigated in this research, hnRNPA1 or FUS, can form functional amyloid fibers in response to cellular stress, but they can also harbor disease-causing mutations. These proteins have a modular architecture that includes one or more nucleic acid binding domains as well as disordered regions that are responsible for their assembly into functional or pathological amyloid structures.
In recent years, the structures of various amyloid fibers formed by fragments of RNPs have been solved. However, these assemblies may not necessarily coincide with those adopted in the context of complete proteins, as is the case of the structure obtained for hnRNPDL-2 solved in our group. In fact, our structure differs significantly from previous ones and questions some of the assumptions that were considered valid regarding the regulation of these proteins in cells.”
Salvador Ventura, Professor and Researcher, Biochemistry and Molecular Biology, Institute of Biotechnology and Biomedicine
Special techniques for resolving functional amyloids
The cryo-EM technique was used by the research group to resolve the structure of hnRNPDL-2 in its assembled state, as well as special techniques for resolving amyloid structures.
This technique has solved a significant number of amyloid fiber structures in the last two years, but these primarily correspond to pathological amyloids involved in systemic and neurodegenerative diseases.
Salvador Ventura says, “Our discovery highlights the power of cryo-EM to study the function of RNPs and the reasons for their link to disease. These proteins have been little studied until now, but they are associated with diseases such as Alzheimer's, muscular dystrophies, cancer, and neurodevelopmental and neuropsychiatric disorders.”
“Thus, our objective now is to take advantage of the experience acquired with this technique to determine the fibrillary states of other functional amyloids and study the effect of mutations, in order to better understand their implications in health and disease,” concludes Salvador Ventura.
The development of this new technology at UAB will enable researchers to take advantage of the newly installed cryo-EM platform at the Alba synchrotron, in which the University is a partner. Solving this type of structure necessitates a significant amount of computational power. The Protein Folding and Conformational Diseases research group at the IBB, led by Salvador Ventura, has recently acquired a powerful computer to perform these calculations.
Source:
Journal reference:
Garcia-Pardo, J., et al. (2023) Cryo-EM structure of hnRNPDL-2 fibrils, a functional amyloid associated with limb-girdle muscular dystrophy D3. Nature Communications. doi.org/10.1038/s41467-023-35854-0.