One of the most basic and amazing processes that occur in both plants and animals is embryogenesis. It is incredible that, within a matter of weeks after fertilization, a single mother egg cell can transform into a living thing with a complex body structure.
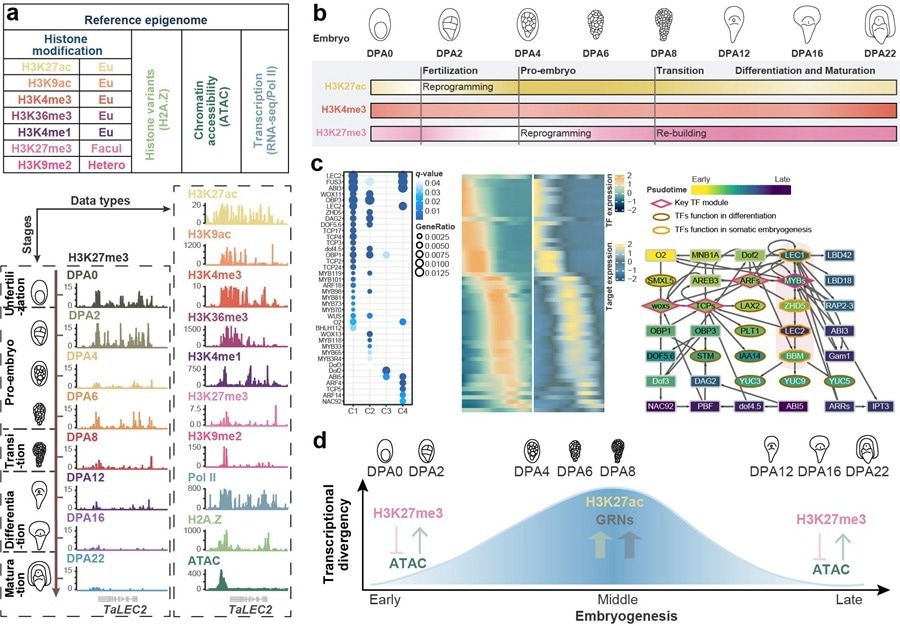 Stage-specific transcriptional divergence regulation model in wheat embryogenesis. Image Credit: IGDB
Stage-specific transcriptional divergence regulation model in wheat embryogenesis. Image Credit: IGDB
Expression of the linked genes and the epigenetic state, which can affect gene expression, have primarily dictated how cells move from one state to another.
In the cellular process of embryogenesis in both plants and animals, there are recognizable and preserved characteristics. Despite the fact that several studies on animal development have been published, gene expression and epigenetic alterations during plant embryogenesis remain mysterious.
Focusing on the allohexaploid wheat, Prof. Jun Xiao’s group at the Institute of Genetics and Developmental Biology (IGDB) of the Chinese Academy of Sciences (CAS) created a “reference epigenome” of wheat embryogenesis.
Seven histone modifications (H3K4me1, H3K4me3, H3K9ac, H3K9me2, H3K27ac, H3K27me3, and H3K36me3), the presence of RNA polymerase II and the histone variation H2A.Z, chromatin accessibility, and transcriptomes were all included in the “reference epigenome.”
The “reference epigenome” assessed the transcription and chromatin state dynamics during embryonic development and offered hints about the regulatory processes that underlie cooperation and competition among the sub genomes of a hexaploid genome.
Their most recent research shows that after fertilization, wheat embryos underwent a significant shift in transcription and chromatin status. At two days after anthesis (DPA2), the histone modification H3K27ac (active gene expression) reduced and largely affected floral genes.
These genes can be silenced and the zygotic cell can be severed from the egg cell by H3K27ac reset. At DPA4, H3K27me3 (which represses gene expression) was decreased and mostly indicated genes relevant to the stem cell niche.
These genes could be activated and cell division facilitated by the H3K27me3 resetting process. This finding implies that zygotic activation (ZGA) in wheat could be a consequence of decreased levels of H3K27ac and H3K27me3.
Even though the cellular mechanism of the maternal to zygotic transition is the same in animals and plants, it is interesting that this epigenetic dynamic pattern was distinct from that in mammals.
In wheat, chromatin accessibility and H3K27ac alteration increased during mid-embryogenesis, resulting in a permissive chromatin environment. This chromatin state allowed transcription factors to bind to the cis-regulatory regions of genes more easily.
The researchers built the gene regulatory networks for embryo patterns based on the cis- and trans-regulation aspects, which might make it easier to analyze the function of the genes. The chromatin once more became condensed in late development, with an increase in H3K27me3.
The differentiation-related genes were also discovered to be surrounded by condensed chromatin, which silenced the genes. This could be the cause of plants’ inability to undergo more complex organogenesis like animals.
Combining early and late embryogenesis, the chromatin state was condensed yet accessible in wheat embryogenesis. According to the sub-genome comparison, this was connected with the expression pattern of similar-different-similar.
The bias expression of homeologs could also be impacted by various epigenetic changes and Transposable element insertion at the three sub-genomes. In comparison to plants with lower genome sizes, like Arabidopsis and rice, wheat has a larger and more complicated distal regulatory system.
This work offers a hitherto unattainable epigenomic resource for wheat embryogenesis research, which can aid in the functional analysis of important genes during embryogenesis, particularly ZGA.
Source:
Journal reference:
Zhao, L., et al. (2023). Dynamic chromatin regulatory programs during embryogenesis of hexaploid wheat. Genome Biology. doi.org/10.1186/s13059-022-02844-2