For cis-regulatory elements in the genome to function, which is crucial for gene regulation, a high-order chromatin structure is required. Three-dimensional (3D) genome organization in eukaryotes exhibits a hierarchical structure. Chromatin can be separated into many structural domains, including chromosomal region, A/B compartment, topologically associated domain (TAD), and chromatin loop.
 Pan-3D genome of TAD boundaries and TE distribution patterns. Image Credit: NI et al.
Pan-3D genome of TAD boundaries and TE distribution patterns. Image Credit: NI et al.
Numerous research has been done on the dynamic alterations to the 3D genome that occur throughout mammalian embryonic development. However, detailed research on the genetic variety of the 3D genome in plants, particularly higher plants, has not been done.
Furthermore, nothing is known about the connection between genomic variation and 3D genomic variation, in addition to the impact of 3D genomic variation on crop domestication.
The pan-3D genome of soybean was created by a research team under the direction of Prof. Zhixi Tian from the Institute of Genetics and Developmental Biology of the Chinese Academy of Sciences. This research demonstrates the intrinsic links between the soybean genome, 3D genome, and gene expression. Genome Biology published the research.
By carrying out high-throughput chromatin conformation capture tests using 27 soybean germplasm materials that were de novo assembled in earlier work, the researchers were able to gather high-quality 3D genome data.
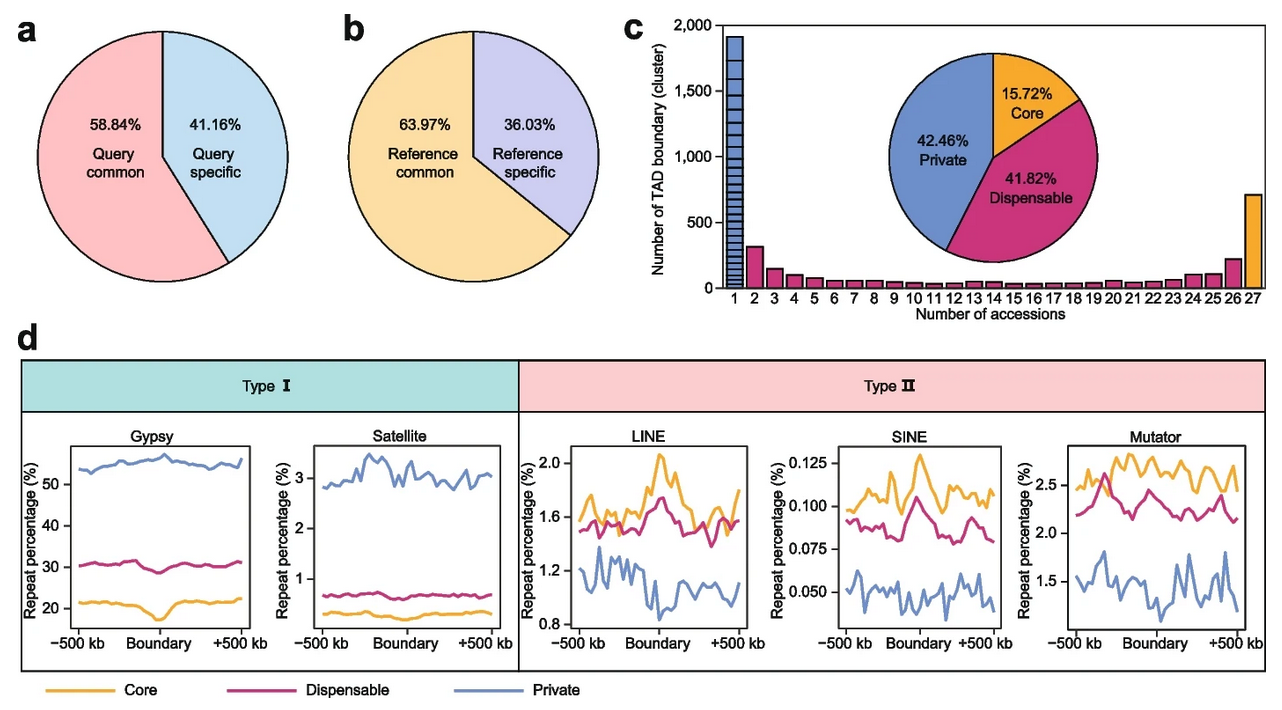
They carried out a pan-omics study and built the pan-3D genome of soybean to look at the conservation and variability of the 3D genome. A/B compartments were largely preserved among soybean accessions, and variations in A/B compartments were strongly associated with genetic traits, the researchers found.
Furthermore, the primary site of A/B compartment transition was found in regions with intermediate genetic characteristics.
The spectrum of interactions between TADs is defined by TAD boundaries. TAD boundary variation frequently indicates a shift in the way genes are regulated. Additionally, the scientists created a pan-3D genome of TAD boundaries. TAD borders displayed more variance than A/B compartments, according to the pan-3D genome.
Further investigation showed that LINE and SINE non-LTR retrotransposons were enriched at TAD boundaries, indicating that these two kinds of elements play significant roles in maintaining TAD boundaries.
Additionally, Gypsy elements and satellite repeats were found to be enriched around accession-specific TAD borders, suggesting that these elements play distinct roles in the development of these boundaries. These findings provide the first explanation of how transposable element (TE) superfamilies alter the 3D genome of plants.
The major source of genetic variation is genomic structural variations (SVs). The link between 3D genomic variation and genomic SV has not been studied in plants due to a paucity of high-quality SV data.
Based on high-quality SV data from de novo genome assembly, the researchers investigated the link between genomic SV and 3D genomic variation in further detail. According to the study, the presence and absence of variation is the most significant contributor to 3D genomic variation.
Further investigation revealed that the SVs that constituted the accession-specific TAD boundaries had much more Gypsy elements and satellite repeats.
These findings demonstrated that TEs can influence SVs to modify 3D genome evolution. They validated the association between 3D genome and gene expression at various levels to investigate the connection between 3D genome diversity and gene expression.
The selection process of the 3D genome in wild soybean, landraces, and cultivars during domestication and improvement was also studied. They discovered that domestication, rather than improvement, was when the 3D genome was most frequently selected. This selection altered the regulation of genes and altered how the soybean gene expression was expressed.
This work investigates the genetic diversity of 3D genome among plant germplasm through pan-3D genome construction, reveals the roles of plant TEs in reshaping 3D genome, analyzes the 3D genome variation caused by genomic SVs, and the selection and subsequent functional effects of 3D genome during crop domestication.”
Zhixi Tian, Study Corresponding Author and Professor, Institute of Genetics and Developmental Biology, Chinese Academy of Sciences
Tian added, “These studies provide a new way to understand the evolution of plant genome, and also provide valuable resources for molecular design breeding.”
Source:
Journal reference:
Ni, L., et al. (2023). Pan-3D genome analysis reveals structural and functional differentiation of soybean genomes. Genome Biology. doi.org/10.1186/s13059-023-02854-8