By carefully constructing upstream open reading frames (uORFs), Caixia Gao’s team at the Institute of Genetics and Developmental Biology of the Chinese Academy of Sciences (CAS) has created a new technique for downregulating gene translation to a predictable and desirable level in plants.
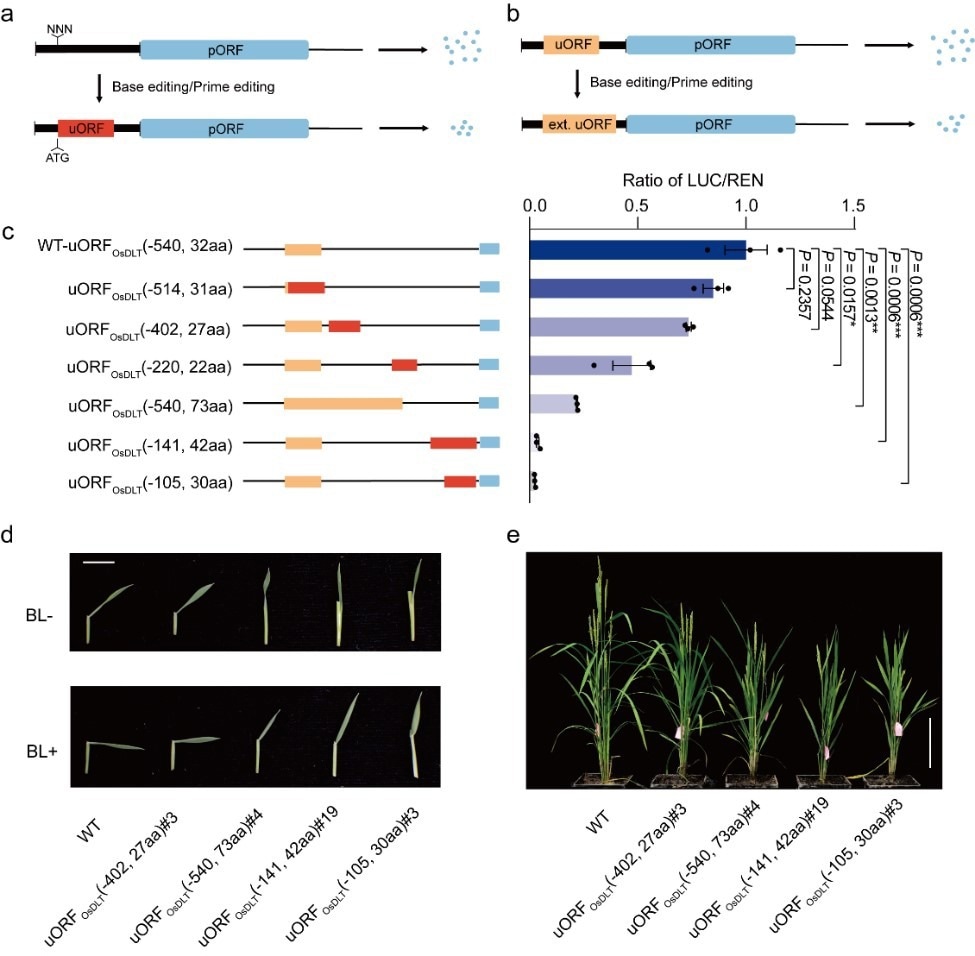 Breeding plants with a predicted phenotype by engineering uORFs. a. Repressed protein expression by generating de novo uORFs; b. Repressed protein expression by extending endogenous uORFs; c. Downregulating protein expression in a graded fashion by producing uORFs with diverse inhibitory activities; d, e. BR sensitivity (d) and morphology (e) of wild type (WT) plants and T1 progenies of edited plants carrying the engineered uORFs. Image Credit: IGDB
Breeding plants with a predicted phenotype by engineering uORFs. a. Repressed protein expression by generating de novo uORFs; b. Repressed protein expression by extending endogenous uORFs; c. Downregulating protein expression in a graded fashion by producing uORFs with diverse inhibitory activities; d, e. BR sensitivity (d) and morphology (e) of wild type (WT) plants and T1 progenies of edited plants carrying the engineered uORFs. Image Credit: IGDB
On March 9th, 2023, the work was published online in Nature Biotechnology.
The development and use of plant genome editing have transformed crop breeding based on molecular design. For breeding new and desired features into crops, techniques for fine-tuning gene expression based on specific genome alterations are essential.
Gene transcription is generally completely prevented by widely used gene editing methods like CRISPR-Cas, CRISPR interference, and RNA interference, or reduced to a specific but unpredictable degree.
A technique for establishing a wide variety of gene expression levels and producing quantifiable phenotypic alterations at the transcriptional level is the CRISPR-Cas9-driven mutagenesis of promoters. Methods for managing gene activity would be further enhanced by a strategy for reliably and gradually modulating endogenous translational gene expression.
By eliminating endogenous uORFs, Gao’s team took the lead in the creation of an effective and controllable technique for upregulating protein expression in 2018. 2020 saw the application of this method to the genetic enhancement of strawberries, leading to the creation of several new strawberry germplasms with various sugar content.
In eukaryotes, uORFs are crucial and ubiquitous cis-regulatory elements that are hypothesized to be related to a reduction in mRNA translation. The inhibitory action of uORFs is influenced by a variety of variables, including uORF length and the distance between uORFs and primary open reading frames (pORFs).
As a result, the researchers suggested two methods for inhibiting gene translation: adding de novo uORFs to a target gene’s 5’ untranslated region (5' UTR) and altering the stop codons of endogenous uORFs to increase their potential to block gene translation.
In transient systems, dual-luciferase and western blot experiments revealed that the newly created and extended uORFs decreased LUC/REN activity ratios to 9.5-86.9% of wildtype levels but had no impact on LUC/REN mRNA levels, suggesting translational control.
The researchers were able to produce mutant rice plants with new and expanded uORFs by using precise genome editing tools like base editors and prime editors. They then verified that these created uORFs had the same impact on phenotypes and protein expression levels as those seen in the transient reporter system.
Using precise genome editing techniques like base editors and prime editors, the researchers were able to create mutant rice plants with new and larger uORFs. They later confirmed that the effects of these newly produced uORFs on phenotypes and protein expression levels were identical to those noted in the transient reporter system.
Additionally, they obtained a variety of plants with different BR sensitivity, plant height, and several tillers by editing the 5’ UTR of rice OsDLT, which encodes for DWARF AND LOW-TILLERING, a member of the GRAS family of transcriptional regulators involved in the brassinosteroid (BR) transduction pathway. This modification matched the corresponding reduction in OsDLT levels seen in a transient protoplast assay.
The researchers have created an effective and broadly applicable strategy for downregulating gene translation to predictable and desirable levels in plants through uORF engineering, which will significantly enhance crop breeding in the future.
Source:
Journal reference:
Xue, C., et al. (2023). Tuning plant phenotypes by precise, graded downregulation of gene expression. Nature Biotechnology. doi.org/10.1038/s41587-023-01707-w