The mesoscopic scale in physics and chemistry relates to the length scale on which the properties of a material or phenomenon can be researched without delving into the behaviour of individual atoms. Because atomic scales are integrated with the continuous scale in a mesoscopic model, they are challenging to construct.
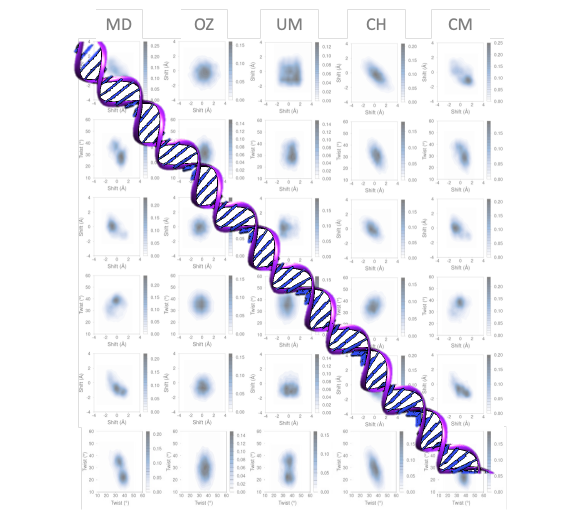
Image Credit: Institute for Research in Biomedicine.
Kim López-Güell, a Maths4Life student, worked with Dr Federica Battistini and Dr Modesto Orozco in the Molecular Modeling and Bioinformatics lab at IRB Barcelona to create a new model of DNA flexibility.
The model created, which incorporates correlation and multimodality at the tetramer level and uses a low-cost computer programme, produces results of exceptional quality. The model is distinguished by its computational efficiency and precision, making it an alternate approach to investigating the dynamics of lengthy DNA segments and opening up the prospect of reaching closer to the chromatin scale.
This work is a milestone in the mesoscopic simulation of DNA. It presents a systematic and comprehensive study of DNA movement correlations and a new method to capture them.”
Dr Federica Battistini, Postdoctoral Researcher, Institute for Research in Biomedicine
This research, carried out in partnership with the “BioExcel” Center of Excellence for Computational Biomolecular Research, contributes to a better understanding of sequencing-dependent DNA at the base pair resolution level.
Several techniques and simplifications have been employed to research this problem throughout the years, but none have resulted in a multimodal model. The suggested method enables for high accuracy local and global descriptions for atomic-level molecular simulations and experimental measurements.
DNA Movement as an Axis
Molecular dynamics is a computational technique that simulates DNA movement, dimeric, trimeric, or tetrameric folding, and even interactions with proteins and drugs. This method allows researchers to analyse activities that occur on time intervals ranging from picoseconds to minutes and apply to molecular systems of varying sizes, making it critical for research into cell functions and disease mechanisms.
The current research describes how DNA moves by employing a low-cost computational technique that predicts the flexibility and conformation of long DNA strands, which might be expanded to RNA duplexes and perhaps long polymers. This new model is likely to help scientists who work with nucleic acid simulations.
Source:
Journal reference:
López-Güell, et al. (2023). Correlated motions in DNA: beyond base-pair step models of DNA flexibility. Nucleic Acids Research. doi.org/10.1093/nar/gkad136.