Anticancer drugs are essential in cancer treatment, but their toxicity is not always restricted to cancer cells, resulting in serious side effects. Scientists are currently focused on molecules that are less harmful to cells to produce anticancer therapies that have fewer side effects on people. The “kinesin inhibitors” are one such class of drugs.
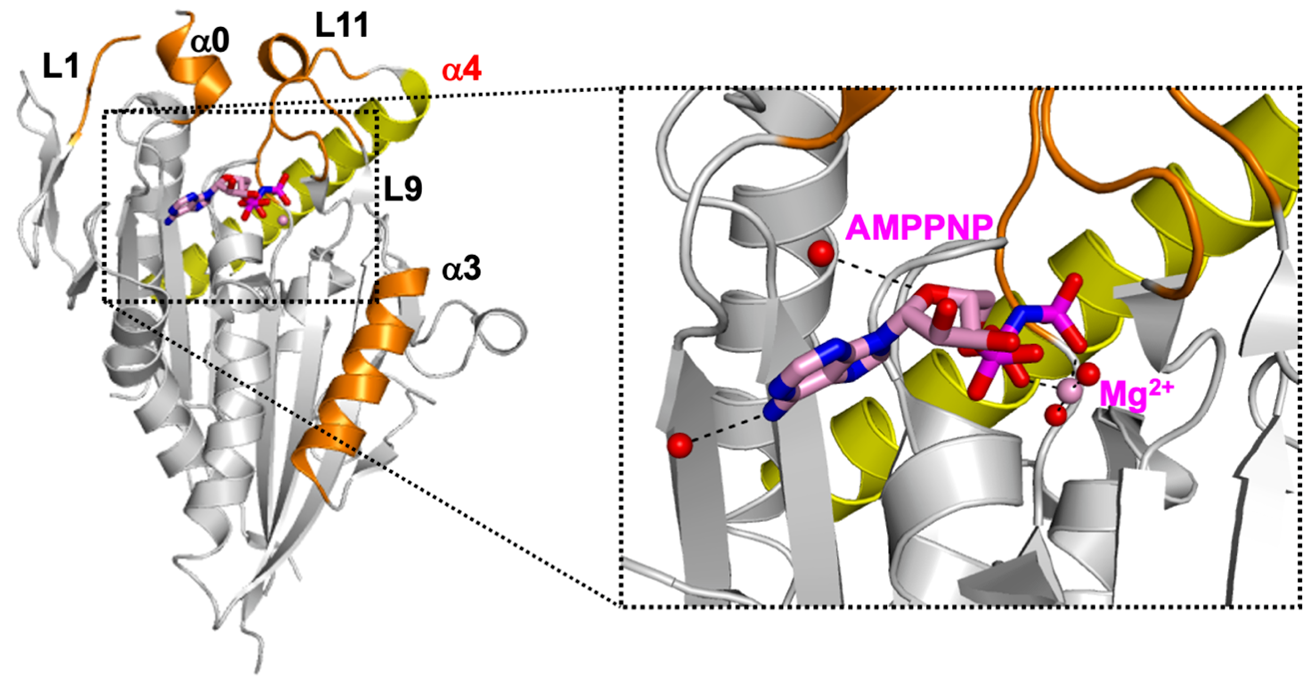 Crystal structure of human CENP-E motor domain—AMPPNP complex. Researchers from Tokyo University of Science unraveled the structure of a complex formed by centromere-associated protein E (CENP-E) motor domain and adenylyl-imidodiphosphate (AMPPNP), a non-hydrolyzable ATP analog. Understanding this crystal structure could help in the development of promising anticancer drugs with fewer side effects. Image Credit: Hideshi Yokoyama from Tokyo University of Science, Japan.
Crystal structure of human CENP-E motor domain—AMPPNP complex. Researchers from Tokyo University of Science unraveled the structure of a complex formed by centromere-associated protein E (CENP-E) motor domain and adenylyl-imidodiphosphate (AMPPNP), a non-hydrolyzable ATP analog. Understanding this crystal structure could help in the development of promising anticancer drugs with fewer side effects. Image Credit: Hideshi Yokoyama from Tokyo University of Science, Japan.
These inhibitors stop cancer progression by specifically targeting kinesin motor proteins, which are necessary for cancer cell division. CENP-E, a member of the kinesin motor protein family, is a viable target for inhibitor therapy because it is required for tumor cell replication. However, identifying the structure of CENP-E is critical to identify inhibitor molecules capable of binding to CENP-E and inhibiting its action.
Surprisingly, binding of the energy molecule ATP to the motor domain of CENP-E modifies its structure or configuration. This also happens when CENP-E attaches to an inhibitor. So far, relatively few CENP-E inhibitors have been reported, and none have received clinical approval. As a result, it is important to collect structural information on the CENP-E motor domain.
To that purpose, a team of researchers from Japan’s Tokyo University of Science (TUS) employed X-Ray crystallography to determine the crystal structure of the complex produced by the CENP-E motor domain and a kinesin inhibitor.
The research, headed by Professor Hideshi Yokoyama of TUS, was published in FEBS Letters on February 23rd, 2023, along with co-authors Ms. Asuka Shibuya of TUS and Assistant Professor Naohisa Ogo, Associate Professor Jun-ichi Sawada, and Professor Akira Asai of the University of Shizuoka.
CENP-E selectively acts on dividing cells, making it a potential new target for anticancer drugs with fewer side-effects.”
Hideshi Yokoyama, Professor, Tokyo University of Science
The CENP-E motor domain was first produced in bacterial cells, then purified and combined with adenylyl-imidodiphosphate (AMPPNP), a non-hydrolyzable ATP analog. To gather comprehensive X-Ray data, the mix was crystallized. The structure of the CENP-E motor domain-AMPPNP complex was determined using these data.
The structure was then compared to that of CENP-E-bound adenosine diphosphate (CENP-E-MgADP) and other commonly known kinesin motor protein-AMPPNP complexes. Based on these observations, the researchers hypothesized that helix alpha 4 in the motor domain was responsible for the loose binding of CENP-E to microtubules, which are cell structures important for cell division.
Compared to the α4 helices of other kinesins, the α4 of CENP-E binds slowly and with lesser strength to microtubules as compared to other kinesins, throughout the ATP hydrolysis cycle.”
Hideshi Yokoyama, Professor, Tokyo University of Science
The uncovering of the complex’s crystal structure is expected to allow more structure-activity connection research, bringing scientists one step closer to designing anticancer drugs that target CENP-E. The research team is hopeful about the future applications of their research and believes that the methodologies used in this research will allow them to design drugs.
The ultimate goal is to use the preparation and crystallization methods described in our study for future drug design studies that aim at developing anticancer drugs with fewer side-effects.”
Hideshi Yokoyama, Professor, Tokyo University of Science
The current research is expected to give cancer patients new hope and help them cope with the negative effects of treatment.
Source:
Journal reference:
Shibuya, A., et al. (2023). Crystal structure of the motor domain of centromere‐associated protein E in complex with a non‐hydrolysable ATP analogue. FEBS Letters. doi.org/10.1002/1873-3468.14602.