Researchers investigating the growth of a roundworm discovered a tiny RNA molecule that controlled the expression of specific genes in the early 1990s. This resulted in the discovery of microRNAs (miRNAs), which are now known to exist in all forms of life. These molecules, it turns out, play vital functions in a variety of biological processes.
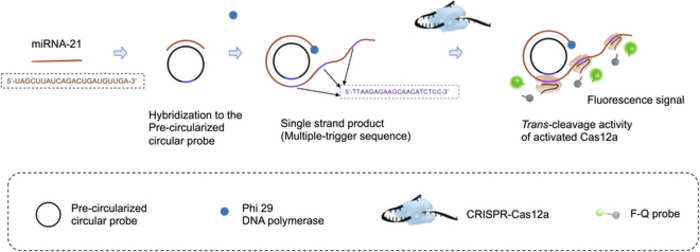
After hybridizing to the precircularized circular probe, the target miRNA is amplified into a long ssDNA product by RCA with massive trigger sequences repeats. The trigger sequence is designed as the activator of Cas12a for showing trans-cleavage activity. Then, the trigger chain is recognized by Cas12a–crRNA duplex and hybridizes to it. Cas12a was then triggered to demonstrate trans-cleavage activity, F-Q probe (an ssDNA probe labeled with a fluorophore at the 5′ end and a quencher at the 3′ end) was cleaved, resulting in the separation of the quencher from the fluorophore, generating the fluorescence signal. Image Credit: BioDesign Research
Researchers discovered a few years later that diseases might dysregulate the production of miRNAs, underlining their potential as biomarkers. Indeed, abnormal miRNA expression is a key characteristic of all tumor-related diseases. Thus, miRNA detection approaches may be effective for cancer screening in the initial stages.
But miRNAs are small and degrade quickly, which makes their rapid identification and estimation difficult. To identify miRNAs in a sample, it is extremely important to first “amplify” them.
Simply said, this entails duplicating a target miRNA several times via biochemical processes so that the miRNA is simpler to detect using low-cost approaches. Unfortunately, most cutting-edge miRNA amplification techniques can take more than five hours to complete, limiting their application in point-of-care testing.
A group led by Associate Professor Chong Zhang of Tsinghua University in China recently pioneered a new technology for fast miRNA amplification and identification. The researchers integrated two well-studied biochemistry procedures into one in a way that considerably reduced the overall time necessary. The study was published on March 27th, 2023, in BioDesign Research journal.
The first method they utilized is known as rolling circle amplification (RCA). The goal of RCA is to create a circular DNA molecule, or “probe,” to which the target RNA fragment attaches. The RNA fragment is then extended by adding nucleotides corresponding to the circular probe after DNA polymerase enzymes and the appropriate DNA building blocks are added. This procedure yields a lengthy, single strand of genetic material containing numerous copies of the circular probe.
This is where CRISPR-Cas12a, the second method, comes into play. CRISPR-Cas12a is a commonly used genetic tool that involves engineering a molecular complex to bind to a specific DNA sequence.
In this case, the complex was designed to attach to a region in the complementary sequence to the circular probe. That is, the CRISPR-Cas12a complexes bound many times along the RCA-produced single-strand of DNA.
When these complexes were bound, the Cas12a component became active, separating a fluorescent probe from its quencher. In turn, this produced an easily recognizable fluorescent signal that was brighter the more the initial target RNA was amplified.
Besides the combination of these approaches, the researchers improved the reaction time of the RCA step by using “precircularized probes.” That is, unlike most normal RCA processes, the circular shape of the probes was determined before the reaction. As Prof. Zhang points out, this sped up the detecting process without sacrificing the system’s performance.
The detection of miRNA could be completed in only 70 min, rather than the usual five hours, with an excellent limit of detection of 8.1 pM and very high specificity.”
Chong Zhang, Associate Professor, Tsinghua University
Overall, the suggested method paints a promising future for miRNA identification and biomarker applications.
Our design improves the efficiency of CRISPR–Cas and RCA-based sensing strategies and shows great potential in lab-based detection and point-of-care testing.”
Chong Zhang, Associate Professor, Tsinghua University
Because the techniques used in this methodology are neither prohibitively expensive nor difficult to execute, widespread implementation of the proposed strategy in clinical settings is possible.
These efforts will pave the path for improved diagnostic tools for cancer and other disorders involving miRNA expression.
Source:
Journal reference:
Niu, C., et al. (2023). Exploring the Trans-Cleavage Activity with Rolling Circle Amplification for Fast Detection of miRNA. Biodesign Research. doi.org/10.34133/bdr.0010.