Reviewed by Danielle Ellis, B.Sc.May 19 2023
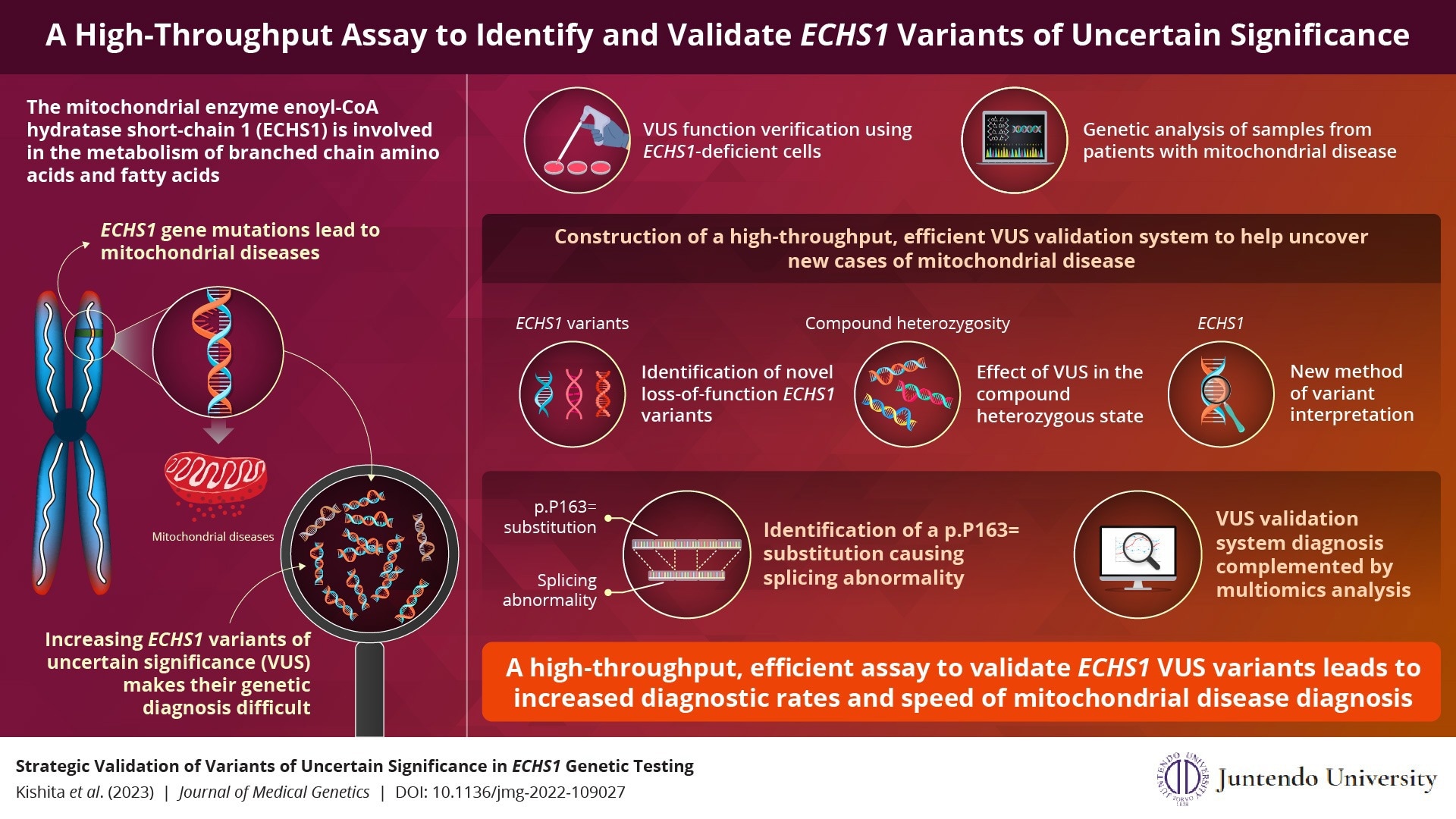
The enzyme variations of mitochondrial enoyl-CoA hydratase short-chain 1 (ECHS1) are dysfunctional. In Japan, they are mostly responsible for mitochondrial disease. Profiling ECHS1 variants of uncertain significance (VUS) is therefore expected to aid in the early diagnosis of mitochondrial disease.
 The assay can speed up mitochondrial disease diagnosis considerably. Image Credit: Yasushi Okazaki from Juntendo University School of Medicine
The assay can speed up mitochondrial disease diagnosis considerably. Image Credit: Yasushi Okazaki from Juntendo University School of Medicine
Researchers in Japan have now created a high-throughput, efficient assay that, when combined with multiomics analysis, can swiftly discover and verify ECHS1 VUS and can also be used to evaluate additional mitochondrial disease genes.
Enoyl-CoA hydratase short-chain 1, a mitochondrial enzyme involved in the metabolism of branched-chain amino acids and fatty acids, is produced by the gene ECHS1. Rare hereditary ECHS1 mutations cause mitochondrial ECHS1 deficiency, which disrupts the metabolism of the vital amino acid valine and causes the buildup of valine intermediates.
In fact, one of the most frequent genes that cause mitochondrial diseases is ECHS1. Along with raising blood lactate levels, these mutant enzymes harm the brain and seriously retard a child’s psychomotor development. The ECHS1 gene has several disease-causing mutations, and ECHS1 variations have been recorded all around the world.
Image Credit: Design_Cells/Shutterstock.com
The identification of several additional instances with ECHS1 mutations has been made possible by recent developments in genetic testing. They have, however, also contributed to a rise in the quantity of ECHS1 variants of uncertain significance (VUS), which provide a significant challenge to genetic diagnosis.
It is common practice to find ECHS1 VUS in malignancies using assays combining Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)/Cas9 and genomic sequencing. However, little is known about how VUS affects uncommon disorders and the molecular processes that give rise to these ECHS1 variants.
Currently, a team of researchers led by Professor Yasushi Okazaki from the Intractable Disease Research Center at Juntendo University in Tokyo, Japan, has created a quick and effective assay technique for verifying ECHS1 variants. The team also includes colleagues Yoshihito Kishita and Ayumu Sugiura.
New research that was released on April 13th, 2023, in The Journal of Medical Genetics describes their results and any possible ramifications they could have.
Diagnosing mitochondrial disease has never been easier thanks to next-generation sequencing technology. However, merely 40% of cases are diagnosed, and 30% have VUS where we cannot predict pathogenicity.”
Yasushi Okazaki, Professor, Division of Diagnostics and Therapeutics of Intractable Diseases, Juntendo University
A high-throughput VUS validation method was necessary because accurate annotation of ECHS1 variants will speed up the genetic diagnosis of mitochondrial diseases and result in the early initiation of treatment.
Cataloging ECHS1 variants by expressing synthetic complementary DNAs (cDNAs) encoding VUS in ECHS1 knockout cells was the initial step in establishing the high-throughput experiment to confirm VUS function.
Using samples from individuals with mitochondrial disease, the team also conducted a genetic analysis. Finally, they used RNA sequencing and proteome analysis to establish the impact on gene expression in confirmed cases.
This functional VUS validation procedure assisted in the discovery of new variants responsible for the loss of the ECHS1 function.
Dr Okazaki added, “Despite its complexity, our high-throughput assay quickly pinpointed the VUS that contributed to the loss of function in ECHS1. Importantly, we identified a new synonymous substitution causing a splicing abnormality, and this assay provides a robust platform to interpret these variants.”
The new technique also has the benefit of being able to detect the impact of compound heterozygosity, which occurs when each parent contributes a mutant allele that harbors a distinct mutation from the other. It also offered a fresh method for variant interpretation.
The group was especially encouraged when their additional “omics” analysis identified several instances that the test had missed. These experts are convinced that by offering a tool to enable the quick genetic identification of early- and late-onset mitochondrial disease, they will be able to enhance the quality of life of patients with mitochondrial disease.
But how important are the study’s findings for future genetic diagnosis?
This validation test can be utilized for the functional assessment of other genes related to mitochondrial diseases, making early treatment a possibility for many individuals.
Dr Okazaki concluded, “The VUS validation of other genes is reproducible and has already begun. Our team has successfully identified variants in the mitochondrial respiratory chain complex I.”
Source:
Journal reference:
Kishita, Y., et al. (2023). Strategic validation of variants of uncertain significance in ECHS1 genetic testing. The Journal of Medical Genetics. doi.org/10.1136/jmg-2022-109027