TEs (transposable elements), particularly LTRs, are recognized to play a crucial role in defining basic genome organization and affecting functional gene expression. Insertion of TE or LTR fragments may also result in the formation of new transcription start sites (TSSs) in the host genome.
 Comparisons of the gene numbers obtained from diploid cotton G. arboreum transcriptomes at different sequencing depths. Image Credit: ©Science China Press
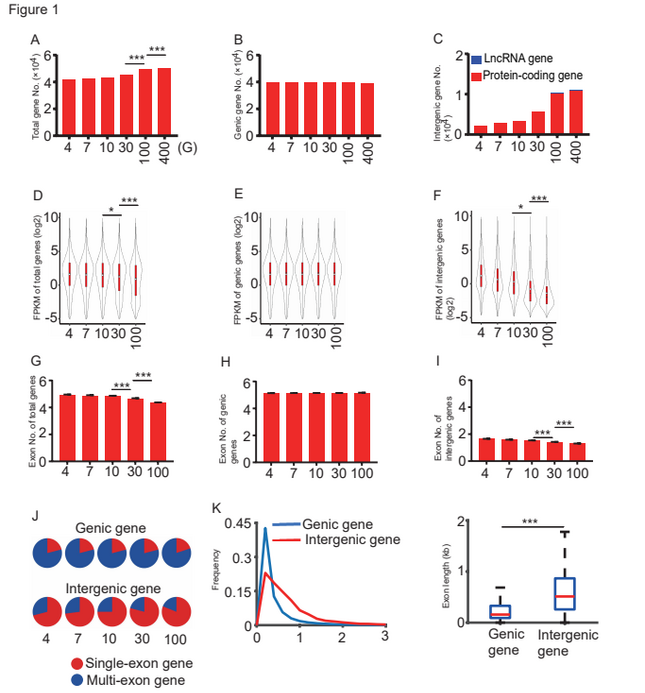
Comparisons of the gene numbers obtained from diploid cotton G. arboreum transcriptomes at different sequencing depths. Image Credit: ©Science China Press
In maize, terminal repeat retrotransposon insertions were assumed to generate new intergenic transcripts by a combination of de novo and homology-based strategies.
These investigations, while predicting the likelihood of novel transcript production via transposon insertion, do not explain the evolutionary, regulatory, or functional processes of these new transcripts. Furthermore, there has yet to be a single systematic research on the extent of intergenic transcript synthesis at the genome level.
Yuxian Zhu and colleagues from Wuhan University’s Institute for Advanced Studies used extraordinarily deep-sequencing techniques (from 10 G to over 100 G) in each cotton sample to identify more than 10,000 unique genes that had not previously been found in genome assembly and annotations. The majority of these transcripts were protein-coding in nature and were produced in diverse ways via LTR insertions.
The researchers discovered that additional transcripts were detected primarily in intergenic areas, as identified in the previously released genome. A total of 10,284 novel intergenic genes were found in the 100 G data collection. There are 10,032 protein-coding genes and 252 lncRNA genes in all.
There was no significant difference in the number of genic genes between these two groups. These new intergenic transcripts were often expressed at extremely low levels, and the majority of them were single exon transcripts.
Due to their low expression level, these novel intergenic transcripts appeared only when the sequencing depth reached 30 G to 100 G. ChIP-seq analysis using antibodies against H3K4me3, H3K27ac, and H3K9me2 demonstrated that the majority of these novel transcripts are unlikely to be transcribed by RNA polymerase II.
Only 30% of these intergenic transcripts had one or two transcription activation signals, whereas more than 70% of genic genes had these markers.
MNase-seq research revealed that genes lacking transcription activation indicators formed their +1 and -1 nucleosomes more closely (approximately only 117 ± 1.4 bp apart), whereas genes with the activation markers had twice as many gaps (around 403.5 ± 46.0 bp apart).
Genes lacking one of these two markers are meant to create -1 nucleosomes near their +1 nucleosomes. This may prevent the RNA polymerase from binding.
According to an evolutionary study, genic genes emerged at 130.8 or 16 MYA during one of the entire genome duplication events, whereas ITG transcripts evolved around 2.3 MYA as a result of the final retrotransposon insertion.
Characterization of these low-transcribed ITG transcripts will aid in the understanding of retrotransposons’ biological involvement throughout speciation and diversification. This research could aid in the understanding of the mechanisms underlying intergenic transcript expression and cotton fiber development.
Source:
Journal reference:
Yang, Y., et al. (2023). Large-scale long terminal repeat insertions produced a significant set of novel transcripts in cotton. Science China Life Sciences. doi.org/10.1007/s11427-022-2341-8.