Urine is one of the common sources for early and sensitive biomarker discovery because it can indicate changes in the human body while still being obtained non-invasively.
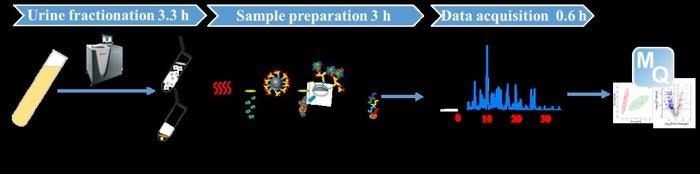 Strategy for Achieving Comprehensive Proteome Analysis of Urine (CPU). Image Credit: The Authors
Strategy for Achieving Comprehensive Proteome Analysis of Urine (CPU). Image Credit: The Authors
But the analysis of the urine proteome presents difficulties as a result of its extensive dynamic range, thereby covering around 10 orders of magnitude in protein concentrations.
The existence of high-abundance proteins in urine could surpass possible disease biomarkers, thereby making their identification complicated.
Generally, fractionation and depletion strategies have been employed before executing mass spectrometry (MS) analysis to improve the detection and identification of such elusive biomarkers.
Considering it as a hopeful alternative, urine fractionation through ultracentrifugation (UC) could be employed to deplete high-abundance proteins, thereby enabling the isolation of exosomes with high purity while keeping the majority of the high-abundance proteins in the supernatant.
Still, the powerful centrifugal shearing force during UC could inevitably interrupt the intact structure of exosomes. This leads to the isolation of just trace amounts from a small urine volume.
Keeping all the current limitations in mind, a research group in China suggested a novel solid-phase alkylation (SPA)-based sample preparation technique for low-loss and anti-interference processing of sub-microgram proteomic samples.
Our method combines UC fractionation, solid-phase extraction (SPA) sample preparation, and liquid chromatography-mass spectrometry (LC-MS) to enable comprehensive proteome profiling of urine, known as CPU, or comprehensive proteome profiling of urine.”
Huiming Yuan, Study Corresponding Author and Professor, Dalian Institute of Chemical Physics, Chinese Academy of Sciences
Yuan added, “This facile strategy resulted in the identification of a total of 1,659 proteins using a short LC gradient of approximately 1 hour, which is 2.3 times more than the 730 proteins identified from raw urine without fractionation.”
Remarkably, in comparison to present urine sample preparation methods, CPU provides considerable benefits. It considerably decreases the analysis time by around 3 to 4 times. and also improves the identification coverage of the urine proteome by almost 130% to 160%.
The method was further employed in combination with label-free quantification to conduct comparative proteome analysis of urine from both IgA nephropathy (IgAN) patients and healthy donors.”
Xinxin Liu, Study Lead Author and Technician, Dalian Institute of Chemical Physics, Chinese Academy of Sciences
Liu added, “As a result, 227 differentially expressed proteins were identified, shedding light on potential biomarkers associated with IgAN.”
Many members of the solute carrier family 22 (SLC22) were found to be up-regulated in IgAN patients. This indicated a possible link to the disruption of renal metabolic function in such individuals.
Our results demonstrated that our developed method holds promise as a valuable tool for the discovery of disease-related biomarkers in urine.”
Xinxin Liu, Study Lead Author and Technician, Dalian Institute of Chemical Physics, Chinese Academy of Sciences
Source:
Journal reference:
Liu, X., et al. (2023) A facile strategy for comprehensive proteome analysis of urine using ultracentrifugation fractionation, solid-phase alkylation based sample preparation and liquid chromatography-mass spectrometry. Urine. doi.org/10.1016/j.urine.2023.05.002.