Reviewed by Danielle Ellis, B.Sc.Jun 20 2023
Aleksandra Radenovic, Head of the School of Engineering’s Laboratory of Nanoscale Biology, has spent years working to enhance nanopore technology, which primarily involves a molecule like DNA going through a tiny pore in a membrane to quantify an ionic current.
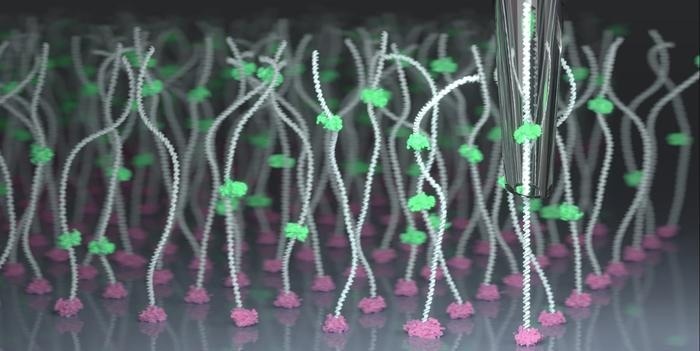 By combining nanopore technology with scanning ion conductance microscopy for the first time, EPFL researchers have achieved near-perfect control over the manipulation of individual molecules, allowing them to be identified and characterized with unprecedented precision. Image Credit: Samuel Leitão/EPFL.
By combining nanopore technology with scanning ion conductance microscopy for the first time, EPFL researchers have achieved near-perfect control over the manipulation of individual molecules, allowing them to be identified and characterized with unprecedented precision. Image Credit: Samuel Leitão/EPFL.
By examining how each nucleotide unnerves this current as it passes through, scientists may discover the sequence of nucleotides in DNA, which encodes genetic information. The findings were reported in Nature Nanotechnology.
Random physical factors influence the movement of molecules via a nanopore and the timing of their analysis, and the fast movement of molecules makes achieving high analytical precision difficult.
Radenovic earlier tackled these difficulties using optical tweezers and viscous liquids. Now, a partnership with Georg Fantner and his team at EPFL’s Laboratory for Bio- and Nano-Instrumentation has resulted in the breakthrough she was searching for, with results that potentially go well beyond DNA.
We have combined the sensitivity of nanopores with the precision of scanning ion conductance microscopy (SICM), allowing us to lock onto specific molecules and locations and control how fast they move. This exquisite control could help fill a big gap in the field.”
Aleksandra Radenovic, Head, Laboratory of Nanoscale Biology, School of Engineering, EPFL
The researchers obtained this control by repurposing a cutting-edge scanning ion conductance microscope built recently at the Lab for Bio- and Nano-Instrumentation.
Improving Sensing Precision by Two Orders of Magnitude
The unanticipated partnership between the labs was facilitated by PhD Student Samuel Leitão. His research focuses on SICM, a technique that leverages fluctuations in the ionic current flowing through a probe tip to generate high-resolution 3D image data.
Leitão used a glass nanopore as a probe to create and apply SICM technology to the imaging of nanoscale cell structures for his PhD. The team used the precision of a SICM probe to move molecules through a nanopore rather than allowing them to diffuse through randomly in this new study.

Image Credit: Dark Gel/Shutterstock.com
The discovery, known as scanning ion conductance spectroscopy (SICS), delays molecule travel through the nanopore, allowing thousands of repeated readings of the same molecule, and even various positions on the molecule, to be taken. The ability to alter transit speed and average several readings of the same molecule has resulted in a two-order-of-magnitude increase in signal-to-noise ratio compared to conventional approaches.
What's particularly exciting is that this increased detection capability with SICS may be transferable to other solid-state and biological nanopore methods, which could significantly improve diagnostic and sequencing applications.”
Samuel Leitão, PhD Student, EPFL
Fantner uses an automotive analogy to summarise the approach’s logic. “Imagine you are watching cars drive back and forth as you stand in front of a window. It’s a lot easier to read their license plate numbers if the cars slow down and drive by repeatedly. We also get to decide if we want to measure 1,000 different molecules each one time or the same molecule 1,000 times, which represents a real paradigm shift in the field,” he adds.
This accuracy and adaptability imply that the method could be implemented to molecules beyond DNA, like protein building blocks named peptides, which could help accelerate proteomics and also biomedical and clinical research.
Finding a solution for sequencing peptides has been a significant challenge due to the complexity of their ‘license plates,’ which are made up of 20 characters (amino acids) as opposed to DNA's four nucleotides. For me, the most exciting hope is that this new control might open an easier path ahead to peptide sequencing.”
Aleksandra Radenovic, Head, Laboratory of Nanoscale Biology, School of Engineering, EPFL
Source:
Journal reference:
Leitao, S. M., et al. (2023). Spatially multiplexed single-molecule translocations through a nanopore at controlled speeds. Nature Nanotechnology. doi.org/10.1038/s41565-023-01412-4.