Reviewed by Danielle Ellis, B.Sc.Jun 27 2023
An improved method of subtyping Alzheimer’s disease based on genetic and tau PET imaging data has been developed computationally. The integrated strategy was successful in finding four subtypes of Alzheimer’s disease and the top genes associated with each, based on a unique clustering framework employing sparse canonical correlation analysis (SCCA).
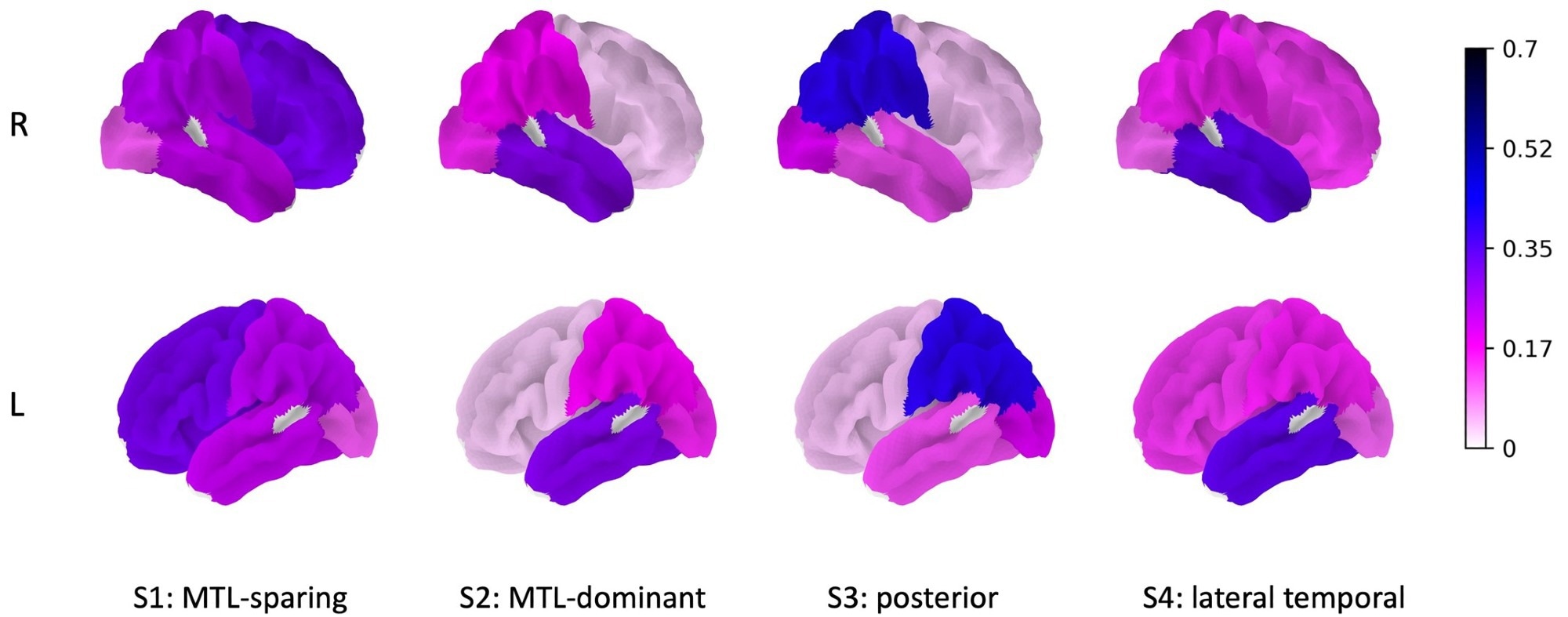 Figure 1. Right and left hemisphere brain surface plots showing the strength of the canonical vector, νƙ, for each subtype. Image Credit: Society of Nuclear Medicine and Molecular Imaging
Figure 1. Right and left hemisphere brain surface plots showing the strength of the canonical vector, νƙ, for each subtype. Image Credit: Society of Nuclear Medicine and Molecular Imaging
The annual meeting of the Society of Nuclear Medicine and Molecular Imaging in 2023 featured a presentation of the finding.
Alzheimer's disease is a complex genetic condition. Based on PET imaging of tau neurofibrillary tangles and amyloid plaques, the pathological indicators of the disease, many subtypes of the condition have been discovered.
Additionally, numerous genes that contribute to the onset of Alzheimer’s disease have been found by recent genome-wide association studies.
By identifying different subtypes of Alzheimer’s disease using both imaging and genomic information, researchers could gain potential new insights into the underlying biology of the disease and its progression. Understanding the specific genetic associations for each subtype could also lead to the development of personalized treatment approaches in the future.”
Joyita Dutta, PhD, Associate Professor, Department of Biomedical Engineering, University of Massachusetts in Amherst
The Alzheimer’s Disease Neuroimaging Initiative (ADNI) imaging and genomics data were used in the study. All ADNI subjects who had undergone 18F-flortaucipir PET and Illumina SNP genotyping (541 total; 334 cognitively normal and 207 cognitively impaired) were included.
Image Credit: Jalisko/Shutterstock.com
Tau PET imaging was used to construct tau PET standardized uptake value ratios from 10 wide regions, and 145 genomic variants linked with Alzheimer’s disease were found and recovered through SNP genotyping. The SCCA-clustering methodology was then used for the tau PET and genomics datasets in collaboration.
The study revealed four subgroups of Alzheimer's disease: medial temporal lobe (MTL)-dominant, posterior, MTL-sparing, and lateral-temporal. Aside from the APOE gene, the top genes related to each subtype were also found.
Dutta added, “Genomics- and imaging-guided individualized subtyping is vital for Alzheimer’s disease because different subtypes may also have distinct rates and profiles of cognitive decline, potentially affecting clinical trial outcomes and treatment response. By combining molecular imaging information with genomics, we have created a diagnostic technique that could be truly personalized for each patient. This has potential for broad diagnostic utility across many disease types, not only Alzheimer’s disease.”