Reviewed by Danielle Ellis, B.Sc.Jun 28 2023
At the Institute of Genetics and Developmental Biology of the Chinese Academy of Sciences, GAO Caixia’s group has introduced the use of artificial intelligence (AI)-assisted techniques to find out novel deaminase proteins with special functions via structural prediction and classification.
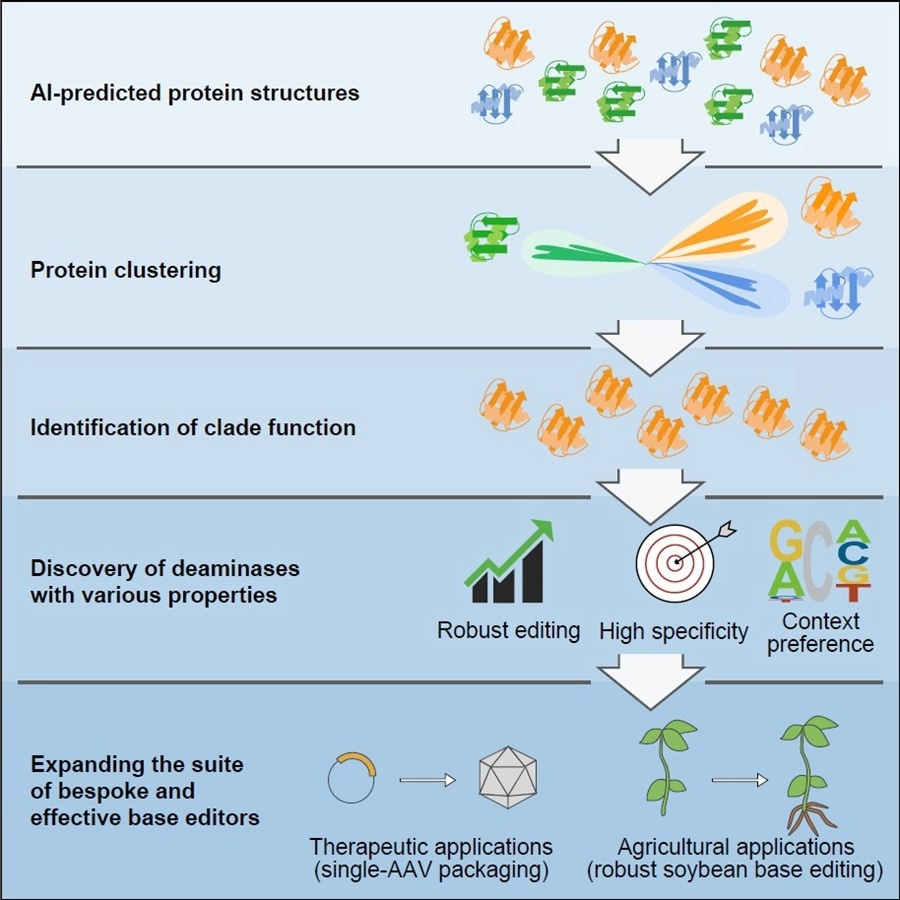 AI-assisted structural predictions and alignments establishes a new protein classification and functional mining method, further enabling the discovery of a suite of single and double-strand cytidine deaminases which show great potential as bespoke base editors for therapeutic or agricultural breeding applications. Image Credit: IGDB.
AI-assisted structural predictions and alignments establishes a new protein classification and functional mining method, further enabling the discovery of a suite of single and double-strand cytidine deaminases which show great potential as bespoke base editors for therapeutic or agricultural breeding applications. Image Credit: IGDB.
This approach has set the stage for a variety of applications for the breakthrough and creation of preferred plant genetic traits.
The study outcomes were reported in the journal Cell.

Image Credit: 3rdtimeluckystudio/Shutterstock.com
The breakthrough of new proteins and the exploitation of various engineered enzymes have added up to the quick advancement of biotechnology. At present, measures to mine novel proteins normally depend on amino acid sequences. It cannot offer a strong link between protein structural information and function.
Base editing is known as a new precision genome editing technology that has the ability to revolutionize molecular crop breeding by initiating preferred traits into elite germplasm. The breakthrough of various deaminases has extended the potential of cytosine base editing.
Even though conventional sequence-based efforts have determined several proteins for use as base editors, restrictions in editing specific DNA sequences or species continue to be the same.
Canonical efforts depend exclusively on protein engineering and directed evolution have assisted to broaden base editing properties, but difficulties are linked. By forecasting the structures of proteins inside the deaminase protein family utilizing AlphaFold2, the scientists clustered and examined deaminases depending on structural similarities.
Around five novel deaminase clusters with cytidine deamination activity in the context of DNA base editors have been determined by the researchers
With the help of this approach, the researchers additionally reclassified a group of cytidine deaminases, known as SCP1.201 and earlier thought to act on dsDNA, to execute deamination mainly on ssDNA.
Through succeeding protein profiling and engineering efforts, the researchers came up with a suite of new DNA base editors with outstanding features. Such deaminases display properties like lower generation of off-target editing events, higher efficiency, editing at different preferred sequence motifs, and much smaller size.
For several therapeutic or agricultural breeding efforts, the scientists stressed that the development of a suite of base editors would allow future tailor-made applications. They came up with the smallest single-strand specific cytidine deaminase, allowing the first efficient cytosine base editor to be packaged in a single adeno-associated virus.
Also, they found a highly effective deaminase from this clade particularly for soybean plants, a globally considerable agricultural crop that earlier displayed poor editing by cytosine base editors.
Normally, the recent emergence of protein structure prediction using increasing genomic databases will considerably expedite the development of new bioengineering tools.
This study stresses a method that makes use of just the cytidine deaminase superfamily to come up with a suite of new technologies and disclose new protein functions. Such newly discovered deaminases rely on AI-assisted structural predictions and considerably extend the utility of base editors for agricultural and therapeutic applications.
Besides, this study will be of huge interest to the larger research community in metagenomics, phylogenetics, genome editing, protein engineering and evolution, and plant breeding.
The study was financially supported by the National Natural Science Foundation of China, the National Key Research and Development Program of China, and the Ministry of Agriculture and Rural Affairs of China, among others.
Source:
Journal reference:
Huang, J., et al. (2023) Discovery of deaminase functions by structure-based protein clustering. Cell. doi.org/10.1016/j.cell.2023.05.041.