A novel technique has been devised by researchers from the University of Leipzig and the University of Vilnius in Lithuania to monitor the smallest twists and torques of molecules in milliseconds. The technique enables the highest resolution real-time tracking of the gene recognition of CRISPR-Cas protein complexes, often known as “gene scissors.”
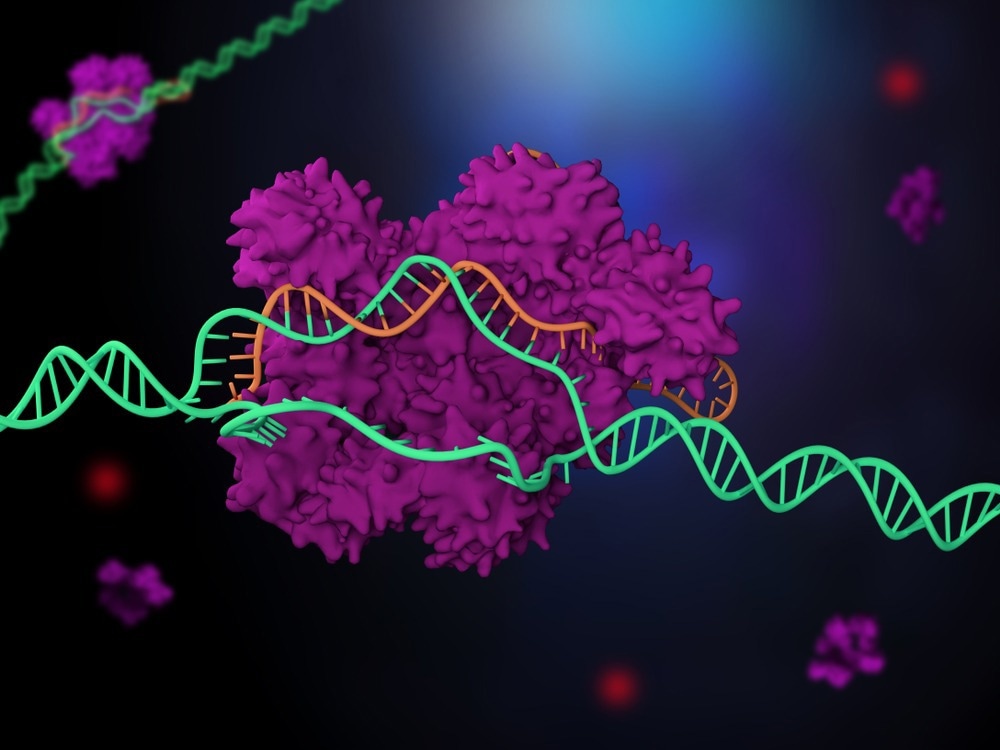
Image Credit: Meletios Verras/Shutterstock.com
It is possible to precisely analyze and simulate the recognition process using the data gathered, increasing the gene scissors’ accuracy. A famous magazine called Nature Structural and Molecular Biology has recently published the findings of the team led by Prof. Dr. Ralf Seidel and Dominik Kauert from the Faculty of Physics and Geosciences.
When a virus attacks bacteria, the bacteria can protect themselves by developing a defense mechanism that repels the virus’s genetic material. CRISPR-Cas protein complexes hold the key to this.
This past decade has seen the discovery and clarification of their role as adaptive immune systems in microbes. The CRISPR complexes identify the invaders through a brief sequence in their DNA and then eliminate them with the aid of an integrated RNA.
Then, specific genes in any organism were turned on and off using the method of sequence recognition with the aid of an RNA. Emmanuelle Charpentier and Jennifer A. Doudna received the 2020 Nobel Prize in Chemistry for their groundbreaking discovery, which transformed genetic engineering.
However, occasionally, CRISPR complexes also respond to gene segments that differ somewhat from the sequence dictated by the RNA. Unwanted side effects occur as a result of medical applications.
The reasons for this are not yet well understood, as the process has not yet been observed directly.
Dominik Kauert, Ph.D. Student, University of Leipzig
Processes At the Nanoscale Followed in Detail
The research led by Prof. Dr. Ralf Seidel and Dominik Kauert makes use of the fact that base pairing with the RNA occurs when the DNA double helix of the target sequence is unfolded during recognition to gain a deeper understanding of the recognition process.
Kauert added, “The central question of the project was therefore whether the unwinding of a piece of DNA that is only 10 nanometers (nm) long can be tracked in real time at all.”
The scientists had to make the unwinding process visible to the microscope to be able to observe it in detail. The scientists employed the arsenal of DNA nanotechnology, which can be used to build any three-dimensional DNA nanostructures, to accomplish this.
The scientists created a 75 nm DNA rotor blade and connected a gold nanoparticle to its end using this so-called DNA origami approach.
In the experiment, the rotation of the gold nanoparticle along a circle with a diameter of 160 nm was translated to the unwinding of the 2 nm thin and 10 nm long DNA sequence; this larger movement could be observed in a particular microscope configuration.
Now that they had this new technique, researchers could almost base pair-by-base pair watch the CRISPR complex Cascade recognize sequences. Surprisingly, only labile binding of the complex occurs during sequence recognition, despite the base pairing with the RNA being hardly energetically beneficial.
There is not a stable link and, as a result, the DNA is damaged until the entire sequence has been fully identified. Process termination occurs if the target sequence is the “wrong” one.
Results Can Help in the Future in the Selection of Suitable RNA Sequences
The researchers have now been able to show that the recognition process’s stochastic character, specifically random molecular movements, is the reason why it occasionally produces false findings.
Kauert noted, “Sequence recognition is driven by thermal fluctuations in base pairing.”
The constructed nanorotors can be employed with other CRISPR-Cas complexes or biomolecules because they are universally effective at sensing twists and torques in single molecules.
Source:
Journal reference:
Kauert, D. J., et al. (2023). The energy landscape for R-loop formation by the CRISPR–Cas Cascade complex. Nature Structural & Molecular Biology. doi.org/10.1038/s41594-023-01019-2