A new study by researchers from the Max Planck Institute for Dynamics and Self-Organization (MPI-DS) found that catalytic molecules can form metabolically active clusters by generating and following concentration gradients.
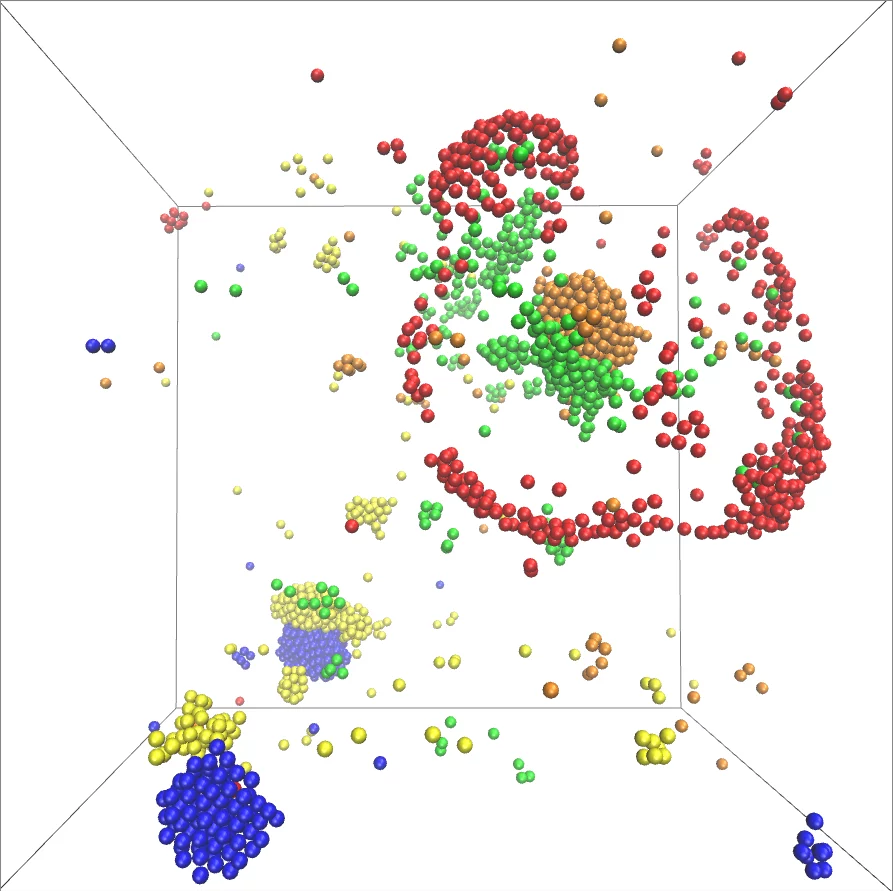 A new model describes the self-organization of catalysts involved in metabolic cycles. Different species of catalysts (represented by different colors) form clusters and can chase each other. Image Credit: © MPI-DS/LMP.
A new model describes the self-organization of catalysts involved in metabolic cycles. Different species of catalysts (represented by different colors) form clusters and can chase each other. Image Credit: © MPI-DS/LMP.
The model suggests the self-organization of molecules in metabolic pathways, potentially introducing a new mechanism to the origin of life theory. The findings may aid in understanding how molecules in complex biological networks can constitute dynamic functional structures, as well as provide a platform for experiments on the origins of life.
The unexpected organization of interacting molecules into cell-like droplets is one possible scenario for the origin of life. The first self-replicating metabolic cycles, which are widespread in biology and present in all organisms, would be created by these molecular species.
The first biomolecules would have to cluster together using slow, ineffective processes, according to this paradigm. Such a slow cluster formation seems inconsistent with the rapid emergence of life.
An alternative model that clarifies such cluster formation and, consequently, the quick onset of the chemical reactions necessary to form life has now been put forth by researchers from the MPI-DS Department of Living Matter Physics.
For this, we considered different molecules, in a simple metabolic cycle, where each species produces a chemical used by the next one. The only elements in the model are the catalytic activity of the molecules, their ability to follow concentration gradients of the chemicals they produce and consume, as well as the information on the order of molecules in the cycle.”
Vincent Ouazan-Reboul, Study First Author, Max Planck Institute for Dynamics and Self-Organization
As a result, the model predicted the formation of catalytic clusters composed of various molecular species. Moreover, cluster growth occurs at an exponential rate. Molecules can thus assemble into dynamic structures very quickly and in large numbers.
Furthermore, the number of molecule species involved in the metabolic cycle influences the structure of the formed clusters.
Our model leads to a plethora of complex scenarios for self-organization and makes specific predictions about functional advantages that arise for odd or even number of participating species. It is remarkable that non-reciprocal interactions as required for our newly proposed scenario are generically present in all metabolic cycles.”
Ramin Golestanian, Director, Max Planck Institute for Dynamics and Self-Organization
Another study discovered that self-attraction is unnecessary for clustering in a small metabolic network. Network effects, on the other hand, can end up causing even self-repelling catalysts to aggregate. The researchers have demonstrated new conditions under which complex interactions can result in self-organized structures.
Overall, the findings of both experiments add another mechanism to the theory of how complex life originated from simple molecules, revealing how catalysts in metabolic networks can form structures in general.