Reviewed by Danielle Ellis, B.Sc.Aug 9 2023
The present understanding of genomic stability and variability is mostly based on a few model species, leaving the scope and generality of these discoveries uncertain. While genome size varies widely across the tree of life, with eukaryotes alone demonstrating a more than 200,000-fold variance, genome size is usually thought to be constant within species or among closely related organisms.
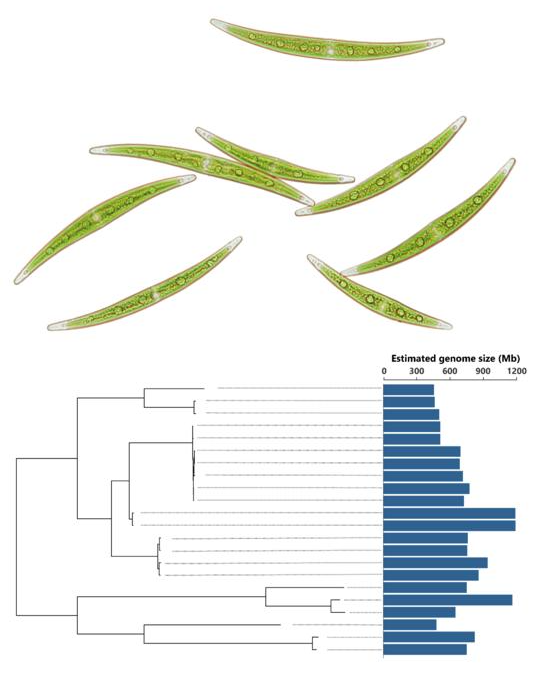 Members of the Closterium peracerosum-strigosum-littorale (C. psl.) complex, a unicellular Zygnematophycean alga, exhibit a more than twofold heritable variation in genome size. A photograph of C. psl. cells taken under the microscope is shown above a phylogeny of C. psl. strains and their associated genome sizes. Image Credit: Yawako Kawaguchi
Members of the Closterium peracerosum-strigosum-littorale (C. psl.) complex, a unicellular Zygnematophycean alga, exhibit a more than twofold heritable variation in genome size. A photograph of C. psl. cells taken under the microscope is shown above a phylogeny of C. psl. strains and their associated genome sizes. Image Credit: Yawako Kawaguchi
However, a recent study by Takashi Tsuchimatsu and Yawako Kawaguchi from The University of Tokyo and their team, published in Genome Biology and Evolution, challenges the conventional notion of genomic stability by revealing striking variation in genome size among the Closterium peracerosum-strigosum-littorale (C. psl.) complex, a group of unicellular algae closely related to land plants.
Tsuchimatsu claims that the study team’s initial plans called for routine population and comparative genomic investigations of 22 naturally occurring C. psl complex strains.
The genomes of Closterium appeared to be much more complex than we had initially thought. Our most exciting finding was that there is extensive genome size variation between closely related algal strains that are morphologically indistinguishable.”
Takashi Tsuchimatsu, Professor, Department of Biological Sciences, Graduate School of Science, University of Tokyo
Surprisingly, the genome sizes of the C. psl. strains in the research varied by more than threefold, ranging from around 450 megabases to over 1,100 megabases. This finding prompted the study’s authors to change the focus of their research to look into the underlying causes of the substantial diversity in genome size.
Tsuchimatsu and colleagues further demonstrated that genome-wide copy number variation (CNV), rather than specific chromosome duplication or proliferation of repeat sequences, plays an essential part in guiding the extensive genome size dynamics observed in this species complex. Genome sequence data were generated from six additional C. psl. strains that differed significantly in the size of their genomes.
Genes or significant DNA regions can be duplicated or deleted, resulting in CNVs. Even among closely related C. psl. strains, the study’s authors discovered that almost 30% of the genes had different copy numbers, which suggests that frequent duplications and deletions that took place across the genome were what caused the fast variations in genome size.

Image Credit: MiniStocker/Shutterstock.com
Additionally, they discovered that for roughly 30% of the genes with CNV, gene expression levels did not rise in direct proportion to gene copy number. This shows that balanced gene expression is maintained despite changes in gene dosage through an epigenetic (i.e., non-Mendelian) mechanism like dosage compensation.
The significant CNV and genome size variation within the C. psl. complex may be preserved through dosage compensation since variations in gene dosage might be harmful.
These findings open up new avenues for investigation into genome dynamics in this species complex and across microeukaryotes more broadly.
Tsuchimatsu added, “Although we found signatures of extensive segmental duplications, we do not yet have a clear map of how duplicate sequences are distributed across chromosomes. For this, it will be necessary to obtain chromosome-scale assemblies together with basic karyotype information including chromosome numbers. Observing chromosomes at high resolution is still tricky in Closterium and this presents a major obstacle.”
But recently, the research team discovered chromosome number variations between C. psl. strains that can mate with one another. This finding suggests a mechanism tolerating chromosomal rearrangements during meiosis (Tsuchikane, et al. 2023) and offers a tantalizing window into additional genome dynamics in this species complex.
The extent of these genomic dynamics could grow clearer with more research in the C. psl. complex and other non-model species. For instance, research by Piganeau and colleagues, which was also recently published in Genome Biology and Evolution, discovered evidence for dosage compensation at the chromosomal level and an unusually high incidence of chromosomal duplications in experimental lines of unicellular green algae.
These “exceptions” thus test the accepted theories of genomic stability by going beyond observations in model organisms, which could eventually show that eukaryotic genomes are significantly more dynamic than previously thought.
Source:
Journal reference:
Kawaguchi, Y. W., et al. (2023). Extensive Copy Number Variation Explains Genome Size Variation in the Unicellular Zygnematophycean Alga, Closterium peracerosum–strigosum–littorale Complex. Genome Biology and Evolution. doi.org/10.1093/gbe/evad115