Reviewed by Danielle Ellis, B.Sc.Aug 30 2023
Markus Buehler of the Massachusetts Institute of Technology integrated attention neural networks, sometimes known as transformers, with graph neural networks to better understand and design proteins in the Journal of Applied Physics from AIP Publishing. The method combines the characteristics of geometric deep learning and language models to not only predict existing protein features but also to imagine novel proteins that nature has not yet created.
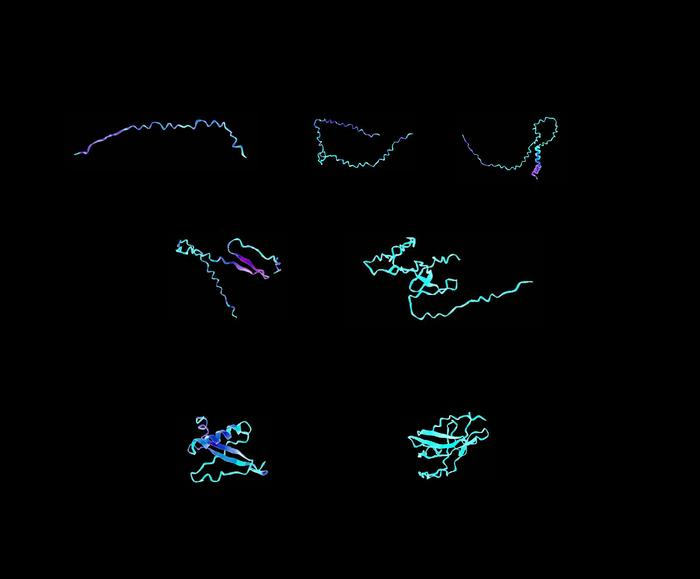 Sample visualizations of designer protein biomaterials, created using a transformer-graph neural network that can understand complex instructions and analyze and design materials from their ultimate building blocks. Image Credit: Markus Buehler
Sample visualizations of designer protein biomaterials, created using a transformer-graph neural network that can understand complex instructions and analyze and design materials from their ultimate building blocks. Image Credit: Markus Buehler
With this new method, we can utilize all that nature has invented as a knowledge basis by modeling the underlying principles. The model recombines these natural building blocks to achieve new functions and solve these types of tasks.”
Markus Buehler, Jerry Mcafee (1940) Professor in Engineering, Massachusetts Institute of Technology
Proteins have been notoriously difficult to simulate due to their complicated architectures, capacity to multitask, and inclination to alter form when dissolved. Machine learning has shown the capacity to transform the nanoscale dynamics that regulate protein activity into functional frameworks. However, going the opposite way—converting a desired function into a protein structure—is still difficult.
Buehler’s technique overcomes this difficulty by converting numbers, descriptions, tasks, and other components into symbols that his neural networks can use.
He began by training his model to predict the sequence, solubility, and amino acid building blocks of various proteins based on their activities. He then trained it to be inventive and construct whole new structures in response to initial parameters for a new protein’s function.
He was able to use this method to make solid forms of antimicrobial proteins that were previously dissolved in water. In another case, his team used a naturally occurring silk protein and developed it into new forms, such as a helix shape for increased elasticity or a pleated structure for increased robustness.
The model accomplished many of the critical duties of building new proteins, but according to Buehler, the technique can integrate additional inputs for additional tasks, possibly making it much more powerful.
“A big surprise element was that the model performed exceptionally well even though it was developed to be able to solve multiple tasks. This is likely because the model learns more by considering diverse tasks. This change means that rather than creating specialized models for specific tasks, researchers can now think broadly in terms of multitask and multimodal models,” Buehler added.
Due to the comprehensive nature of this technique, this model can be used in many fields other than protein design.
Source:
Journal reference:
Buehler, M. J. (2023). Generative pretrained autoregressive transformer graph neural network applied to the analysis and discovery of novel proteins. Journal of Applied Physics. doi.org/10.1063/5.0157367.