Groundbreaking research led by a global group of over 100 researchers will enable a more in-depth exploration of human genetic variation as fully sequencing the Y chromosome, a feat that has challenged scientists for years, has been accomplished for the first time. In this interview, we speak to Dylan Taylor about this impactful research and how it may shape our understanding of human genetics.
Please could you introduce yourself and your current research activities?
I am Dylan Taylor, a Ph.D. candidate and NIH F31 fellow in the Department of Biology at Johns Hopkins University. My work with the T2T consortium focuses on exploring how a complete reference genome can improve our ability to study human genetic variation and how it impacts human traits and health.
How did you become involved in researching human genetic variation?
I initially joined the T2T consortium in 2021 as part of the team exploring the improvements to genetic analyses afforded by using a complete human reference genome. This initial version of the reference genome—published in 2022—while a complete human genome, was from a sample that lacked a Y chromosome. Building on the initial reference genome, I led similar analyses of the now complete human Y chromosome sequence.
Alongside a team of more than 100 researchers, you have fully sequenced the Y chromosome, T2T-Y, for the first time. How does it feel to have contributed to such an important development in human genetics?
It has truly been an honor to be a part of such an amazing team of researchers; I never imagined I would have the opportunity to work on something so impactful. Prior to this, I had never been a part of a collaboration so large, but everything ran very smoothly. The different areas of expertise everyone brought to the table really made this endeavor possible.
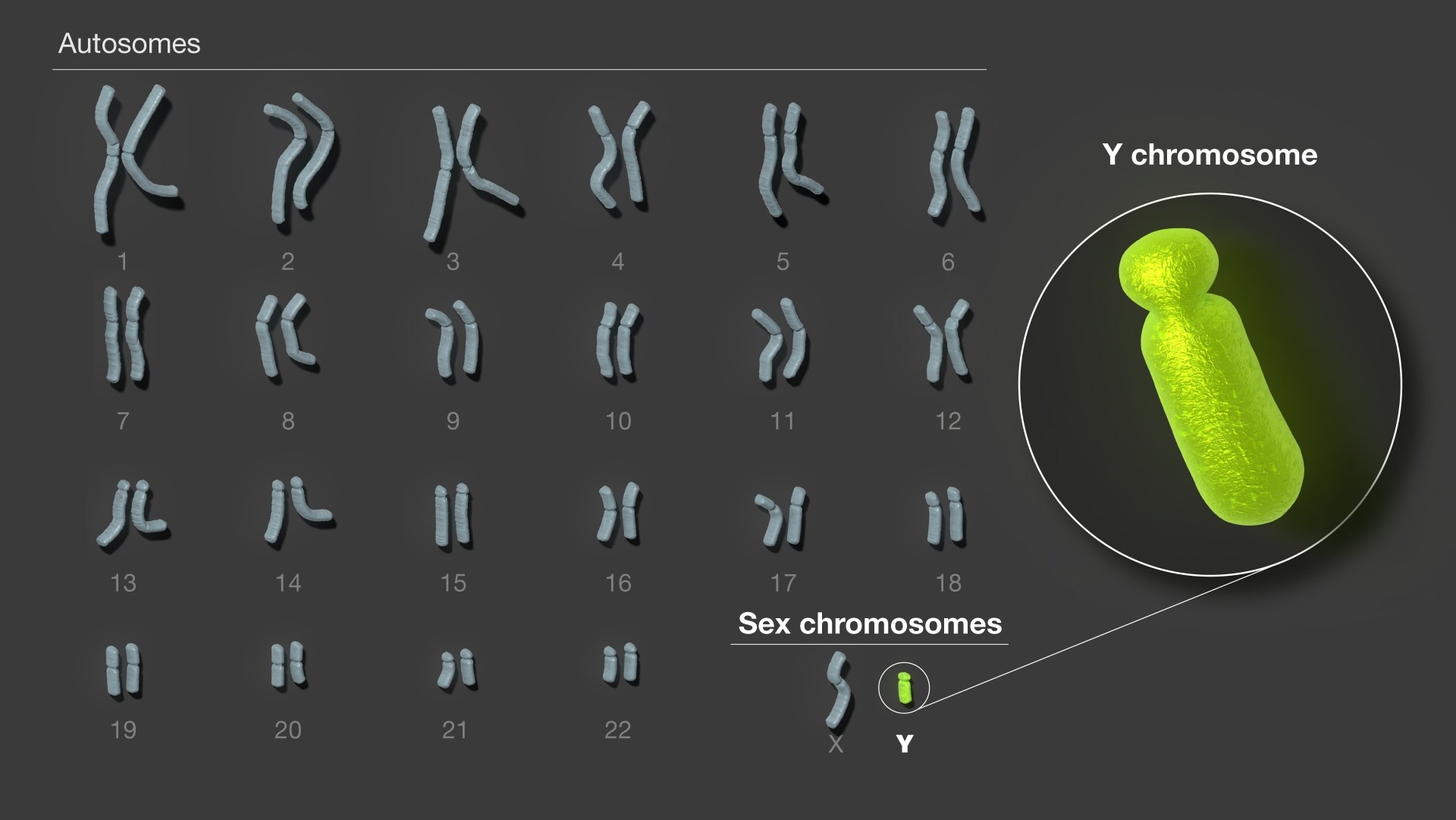
Image Credit: NIH/NHGRI
What are the immediate implications of this research?
As with completing the rest of the human genome, one of the biggest impacts of completing the Y chromosome is that now we can actually dig deeper into those regions that were previously missing/unresolved and figure out what’s going on there. We didn’t know if there were any interesting genetic features in those regions affecting human development and disease because we couldn’t even study them. Now, we have the tools to do just that.
Many may be surprised to hear that we were missing this reference piece of the human genome. Why was a complete sequence of the Y chromosome difficult to assemble?
While it would be terrific if we could sequence an entire chromosome all at once and without any errors, our current genome sequencing technology does not allow that. Instead, we can only sequence short fragments of DNA (called reads) at a time—usually just a couple hundred letters at once.
For reference, the Y chromosome is over 60 million letters long. After sequencing, we have to figure out how those pieces fit together using overlaps between them, almost like doing a jigsaw puzzle. When there are long stretches of repetitive DNA, it becomes very hard to figure out how the pieces fit together.
The hardest parts of a jigsaw puzzle are often places where the pieces are all very similar to each other, like a stretch of blue sky for example. Likewise, it is also very hard to assemble parts of a chromosome that are very similar to each other, and the Y chromosome is filled with these types of sequences.
How did the team approach sequencing the chromosome?
The technology that allowed us to complete the Y chromosome—called long-read sequencing—allows us to sequence fragments of DNA that are much longer: up to hundreds of thousands of letters long. This is sort of like having a bunch of the pieces in your jigsaw puzzle pre-assembled, making it much easier to figure out what those repetitive regions look like.
That said, even with the advantages that long-read sequencing provides, there were whole teams of people focused on developing algorithms to put together the most challenging regions of the chromosome, and manually checking that everything was assembled correctly afterward.
Were there any regions of T2T-Y that were unexpected?
In adding so much sequence that was previously missing, we discovered 41 additional genes on the Y chromosome, many of which are part of gene families known to be involved in sperm regulation. We were also able to fully resolve the structures of other genes thought to play roles in development and fertility.
How does T2T-Y compare to existing reference genomes? Will this change how researchers approach investigations involving the Y Chromosome?
The previous reference genome was missing over half of the sequence of the Y chromosome. Because of the errors and missing sequences in the previous Y chromosome reference, scientists have had a difficult time fully exploring the functions of the chromosome due to its incompleteness. Indeed, it is often left out of genetic studies altogether.
Our hope is that, by revealing the full sequence of the Y chromosome, researchers will be able to dig deeper into those regions that were previously missing or incorrect and figure out what role they might be playing in human biology.
Image Credit: vchal/Shutterstock.com
What are the next steps for this research?
Now that the full sequence of the Y chromosome has been revealed, future research will focus on determining how the Y chromosome functions, and its role in human development and disease. Along with the rest of the complete human genome released last year, this work has laid the groundwork for how to generate more of these high-quality genome assemblies, including complete T2T genomes from many different individuals.
By generating complete genomes from people around the world and those with different diseases, we can more fully understand and explore the scope of human genetic diversity and, therefore, better inform human health research. Right now, at JHU we are using these technologies to explore the genetic basis of pancreatic disease. We are also extending this work beyond humans: completing the genomes of several primate species, allowing us to more accurately explore the evolutionary history of these species and how it relates to human evolution.
Where can our readers go to stay up to date with your research activities?
You can learn more about the T2T consortium and stay up to date on our work at the T2T home page. If you are interested in learning more about my research beyond the T2T consortium, you are welcome to visit my personal website.
About Dylan Taylor
 Dylan Taylor is a Ph.D. candidate in the Cell, Molecular, Developmental Biology and Biophysics (CMDB) program at Johns Hopkins University, and an NIH NRSA/F31 fellow through the National Human Genome Research Institute. He is advised by Dr. Rajiv McCoy.
Dylan Taylor is a Ph.D. candidate in the Cell, Molecular, Developmental Biology and Biophysics (CMDB) program at Johns Hopkins University, and an NIH NRSA/F31 fellow through the National Human Genome Research Institute. He is advised by Dr. Rajiv McCoy.
His research focuses on developing tools and datasets that facilitate deeper insight into human genetic variation. As a member of the T2T Consortium, he has explored the utility of a complete reference genome in studying human genetic variation. His own research focuses on generating large, globally diverse human transcriptomics datasets, and developing computational and statistical methods that use these datasets to investigate genetic variation underlying gene expression and splicing differences between individuals.
He received his B.S. in Biology from the University of Maryland, College Park in 2018.