Reviewed by Danielle Ellis, B.Sc.Sep 20 2023
The human body heals wounds or detoxifies harmful substances like histamine with the help of a group of enzymes called copper amine oxidases. These enzymes play a crucial role, but pinpointing the precise positions of the tiniest hydrogen atoms within them has been a challenge with conventional techniques. However, this knowledge is essential for enhancing these enzymes to exhibit unique and valuable biochemical reactions.
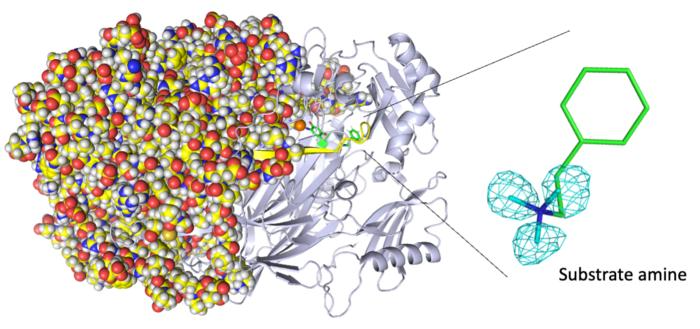 Entire neutron crystal structure of copper amine oxidase. (Left) Neutron crystallography determines the entire protein structure, including the hydrogen atoms. (Right) The precise positions of the hydrogen atoms indicate that the detected molecule is not a product aldehyde but a substrate amine. Maps for the hydrogen atoms (more precisely, deuterium) are presented by using a cyan mesh. Image Credit: Takeshi Murakawa, Toshihide Okajima.
Entire neutron crystal structure of copper amine oxidase. (Left) Neutron crystallography determines the entire protein structure, including the hydrogen atoms. (Right) The precise positions of the hydrogen atoms indicate that the detected molecule is not a product aldehyde but a substrate amine. Maps for the hydrogen atoms (more precisely, deuterium) are presented by using a cyan mesh. Image Credit: Takeshi Murakawa, Toshihide Okajima.
In a recent study published in ACS Catalysis, a team led by researchers from Osaka Medical and Pharmaceutical University and Osaka University used a technique called neutron crystallography to create a detailed, atom-by-atom image of a copper amine oxidase enzyme. This breakthrough study provides unprecedented insights into the enzyme’s biochemistry, shedding new light on its inner workings.
Some copper amine oxidase enzymes display unusual biochemistry, including phenomena like quantum tunneling, which enables exceptionally rapid reaction rates that are otherwise hard to explain. Although pinpointing the precise location of each hydrogen atom within these enzymes can be challenging, it is a critical piece of knowledge when designing artificial enzymes that mimic their behavior.
Image Credit: Design_Cells/Shutterstock.com
Researchers typically determine the atom-by-atom structure of enzymes using X-Ray crystallography. However, this technique relies on the diffraction of electrons within the enzyme and is insufficient for imaging hydrogen atoms, which usually contain only one electron.
In their study, the researchers opted for an alternative imaging technique known as neutron crystallography. This method analyzes diffraction patterns resulting from atomic nuclei in the enzyme, and since all atoms possess atomic nuclei, it allows for the imaging of hydrogen atoms and provides a more comprehensive understanding of the enzyme's structure.
There are pH-dependence, conformational change, and radical intermediate stabilization questions of our enzyme that X-ray crystallography in itself cannot fully explain. Neutron crystallography is well-suited for answering these questions.”
Takeshi Murakawa, Study Lead Author, Osaka University
The researchers gained several valuable insights from their study. Firstly, they were able to visualize the protonation and deprotonation state of specific sites within the enzyme, which is closely related to pH levels. These sites are crucial for stabilizing radical species, which are highly reactive atoms containing an unpaired electron.
Additionally, the study provided insights into the movements of the enzyme's topaquinone cofactor. This cofactor exhibited sliding, upward tilting, and half-rotation motions, all of which play a role in facilitating single-electron transfer processes within the enzyme. These findings enhance our understanding of how the enzyme functions at the atomic level.
We disclose binding of a second molecule of high-affinity amine substrate during the enzymatic reaction, a previously unknown event in the enzyme active site. X-ray crystallography misses such insights.”
Toshihide Okajima, Study Senior Author, Osaka University
This research has uncovered previously unknown structural information about a copper amine oxidase enzyme, which plays multiple roles in biochemical metabolism. Understanding the precise locations of hydrogen atoms within the enzyme is key to explaining why it functions efficiently under normal physiological conditions of temperature and pressure.
In the future, scientists may use these newfound insights to design artificial enzymes with applications in the chemical industry. This knowledge can help create more efficient and tailored enzymes for various industrial processes.
Source:
Journal reference:
Murakawa, T., et al. (2023). Neutron Crystallography of a Semiquinone Radical Intermediate of Copper Amine Oxidase Reveals a Substrate-Assisted Conformational Change of the Peptidyl Quinone Cofactor. ACS Catalysis. doi.org/10.1021/acscatal.3c02629.