Reviewed by Danielle Ellis, B.Sc.Sep 25 2023
A Recent study conducted by Professor Qin Yuan's team from the Center for Genomics at the Haixia Institute of Science and Technology (Future Technology College) at Fujian Agriculture and Forestry University has been published in the prestigious journal Horticulture Research, which is a leading publication in the field of horticultural science.
 Genome size estimation and de novo genome assembly of S. glauca. Image Credit: Horticulture Research.
Genome size estimation and de novo genome assembly of S. glauca. Image Credit: Horticulture Research.
The salinity of the soil is a growing issue for the sustainability of human development and global crop production. To increase crop tolerance to salt stress, it is essential to understand salt tolerance mechanisms and recognize salt-tolerant genes.
S. glauca, a halophyte species that thrives in saltwater environments, has the unusual ability to absorb and hold onto high salt concentrations inside of its cells, particularly in its leaves, which raises the possibility that it has a unique mechanism for tolerating salt.
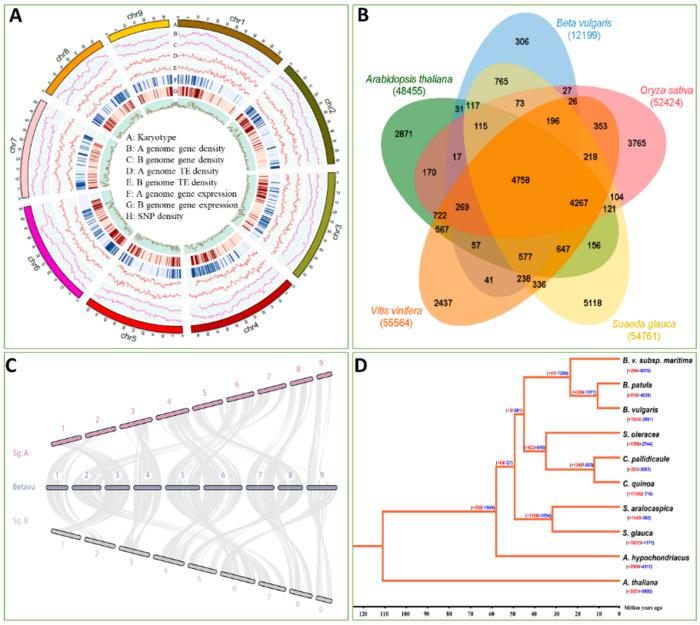
The S. glauca genome was de novo sequenced by the researchers for this study. A total of 54,761 annotated genes, including alleles and repeats, are found in the 1.02 Gb genome, which is made up of two sets of haplotypes. The genomes of S. glauca and B. vulgaris exhibit significant synteny, according to comparative genomic analysis. Retroelements are the most prevalent repeat sequences, making up 70.56% of the S. glauca genome.
Researchers examined genome-wide allele-specific expression in the analyzed samples by utilizing the allele-aware assembly of the S. glauca genome. The findings suggested that consistent allele-specific expression might be influenced by the diversity of promoter sequences.

Image Credit: youli zhao/Shutterstock.com
Sepal, petal, stamen, and carpel are the four whorls that make up the typical angiosperm flower, though some species may have different arrangements. Only three whorls were seen in S. glauca, with the first whorl being severely damaged and the second whorl having sepal-like traits. The typical phenotype can be seen in the third and fourth whorls.
Researchers examined the expression of genes linked to photosynthesis and pathways to chlorophyll synthesis in order to verify the sepal-like nature of the second whorl of the S. glauca flower. The clustered heat maps demonstrated that the second whorl's gene expression patterns matched those of the sepals.
Additionally, a thorough examination of the ABCE gene family shed light on the development of the flower morphology of S. glauca, indicating that the absence of petals in S. glauca is caused by the dysfunction of A-class genes.
Researchers carried out a comprehensive genome analysis between Suaeda species, including Suaeda aralocaspica (a salt-tolerant plant in the Amaranthaceae family) and other non-salt-tolerant species in the same family, to gain a comprehensive understanding of the salt tolerance mechanisms in Suaeda genus plants.
The findings showed that in these two salt-tolerant Suaeda plants, there was a big increase in the number of gene families, specifically 1840 genes. While analyzing these genes more closely, the researchers discovered that they were enriched with functions related to DNA repair, keeping chromosomes stable, removing chemical marks from DNA (DNA demethylation), binding to positive ions (cation binding), and responding to red and far-red light signals.
These functions were particularly common in the gene families that were expanded and shared among different Suaeda species.
When Suaeda species was compared to non-halophytic (non-salt-tolerant) species in the Amaranthaceae family, the researchers found that the expanded genes in Suaeda plants were mostly focused on maintaining the stability of DNA and chromatin. This suggests that the function related to “DNA/chromatin stability maintenance” likely plays a crucial role in enabling Suaeda plants to tolerate salt.
To further confirm the earlier observations, a temporal RNA-seq analysis was conducted on salt-treated Suaeda plants. Notably, a group of genes that consistently increased their activity, referred to as “transition-up genes,” was identified as a stable response mechanism in S. glauca for coping with salt stress.
Interestingly, in the leaves, these transition upregulated genes were primarily associated with functions related to DNA repair and chromosome stability. Additionally, there was an enrichment of genes associated with lipid biosynthesis and isoprenoid metabolic processes, as indicated by GO enrichment analysis.
These findings align with the earlier analysis of the expanded gene families in S. glauca. A comprehensive analysis of transcription factors across the genome revealed a significant increase in the FAR1 gene family. However, additional research is required to fully understand the precise role of the FAR1 gene family in the salt tolerance of S. glauca.
In this investigation, the chromosomal level of the S. glauca haploid genome was de novo assembled. It has been established as a reference genome for the halophyte S. glauca due to the high quality of the genome sequence and the extensive annotation.
Researchers also identified a novel salt tolerance mechanism in halophytic plants called “DNA/chromatin stability maintenance.” This mechanism of salt tolerance in halophytic plants has only recently been discovered, and it may hold new information and opportunities for improving crop salt tolerance.
Source:
Journal reference:
Cheng, Y., et al. (2023). Chromosome-scale genome sequence of Suaeda glauca sheds light on salt stress tolerance in halophytes. Horticulture Research. doi.org/10.1093/hr/uhad161.