Researchers from Hiroshima University have developed a groundbreaking software tool known as DANGER (Deleterious and ANticipatable Guides Evaluated by RNA-sequencing) analysis, offering a transformative solution for safer and more precise genome editing in all organisms with a transcriptome.

Image Credit: Yurchanka Siarhei/Shutterstock.com
CRISPR technology has revolutionized genome editing for the past decade, but it is not without its challenges.
By using DANGER analysis, the research team hopes that these challenges can be overcome, empowering researchers to perform precise on- and off-target assessments without relying on a reference genome, with significant implications for medicine, agriculture, and biological research.
Revolutionizing Genome Editing Safety
Genome editing, or gene editing, is a powerful technology that allows researchers to modify the DNA of an organism, enabling the addition, removal, or alteration of genetic material within the genome.
CRISPR-Cas9, a widely recognized gene editing technology, has gained popularity for its speed, cost-effectiveness, and accuracy. However, CRISPR-based gene editing faces significant challenges, which are briefly discussed below:
Quantitative Monitoring of Phenotypic Effects
One of the key challenges is the quantitative assessment of phenotypic effects resulting from unpredictable CRISPR dynamics. While CRISPR-Cas9 is remarkably accurate, it can still introduce unexpected changes in the DNA sequence, commonly referred to as off-target effects. These off-target effects can lead to unintended alterations in gene expression and undesired phenotypic changes.
Dependence on Reference Genomes
CRISPR technology typically relies on reference genomes as templates for guiding the editing process. The reference genome provides a blueprint of the organism's genetic makeup, helping researchers design their genome editing strategies. However, the challenge arises when dealing with organisms, patients, cancers, or uncharacterized species for which genomic information is incomplete or unavailable. These incomplete or unique genomes do not align with existing reference genomes, making the off-target effects even more unpredictable.
Dr. Kazuki Nakamae, an Assistant Professor at the PtBio Collaborative Research Laboratory at the Genome Editing Innovation Center, Hiroshima University, highlights this issue, stating, "The design of genome editing requires a well-characterized genomic sequence. However, the genomic information of patients, cancers, and uncharacterized organisms is often incomplete."
In response to these challenges, a dedicated research team embarked on developing the DANGER analysis software, a pioneering solution for safer genome editing.
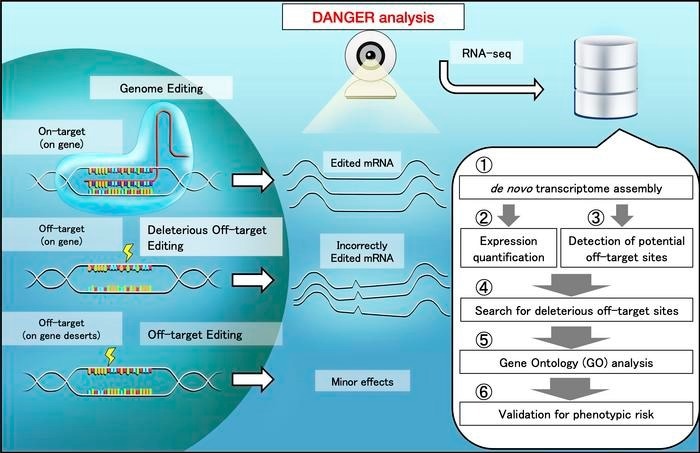 Scheme of safety evaluation by DANGER analysis. When off-target regions are affected by genome editing, unexpected changes in the mRNA quantity and sequence can emerge. The DANGER analysis is designed to analyze the effects from RNA-seq data at the Gene Ontology (GO) level. Credit: Kazuki Nakamae and Hidemasa Bono, Hiroshima University
Scheme of safety evaluation by DANGER analysis. When off-target regions are affected by genome editing, unexpected changes in the mRNA quantity and sequence can emerge. The DANGER analysis is designed to analyze the effects from RNA-seq data at the Gene Ontology (GO) level. Credit: Kazuki Nakamae and Hidemasa Bono, Hiroshima University
Introducing DANGER Analysis
The DANGER analysis is a sophisticated software pipeline that addresses the issues of phenotypic effects and the dependency on reference genomes in genome editing. The researchers conducted extensive work with gene-edited samples of human cells and zebrafish brains, applying risk-averse on- and off-target assessments using RNA-sequencing data.
This innovative analysis pipeline accomplishes several critical goals:
- Detection of On- and Off-Target Sites: DANGER analysis identifies potential DNA on- and off-target sites within the mRNA-transcribed region of the genome using RNA-sequencing data. This allows researchers to pinpoint the exact locations of their edits.
- Evaluation of Phenotypic Effects: The software evaluates phenotypic effects by identifying deleterious off-target sites based on the evidence provided by changes in gene expression. This enables researchers to assess the functional impact of off-target edits.
- Quantification of Phenotypic Risk: Perhaps most impressively, DANGER analysis quantifies phenotypic risk at the gene ontology term level without needing a reference genome. This groundbreaking feature enables the assessment of genome editing in a wide range of organisms, personalized human genomes, and genomes altered by diseases and viruses.
Dr. Hidemasa Bono, a Professor at the Genome Editing Innovation Center, Hiroshima University, expresses the significance of their work:
Our DANGER analysis is a novel software that enables quantifying phenotypic effects caused by estimated off-target. Furthermore, our tool uses de novo transcriptome assembly whose sequences can be built from RNA-sequencing data of treated samples without a reference genome."
Dr. Hidemasa Bono, Professor, Genome Editing Innovation Center, Hiroshima University
Industry Relevance of DANGER Analysis
With its ability to identify on- and off-target sites, evaluate phenotypic effects, and quantify phenotypic risk without reliance on a reference genome, DANGER analysis opens new frontiers in medicine, agriculture, and biological research.
In medicine, DANGER analysis could contribute to safer and more precise genome editing, improving the accuracy and predictability of gene therapy and personalized medicine.
For biological research, DANGER analysis could help researchers explore the genomes of uncharacterized organisms and diseases more effectively. This enables in-depth studies, including the identification of novel genes and functions, with a higher degree of precision.
Looking elsewhere, the new tool may have applications in agriculture. For example, genome editing of crops using DNAGER analysis could allow for the creation of a food source that reliably and accurately meets our national and climate-conscious expectations.
Source:
Journal reference:
Nakamae, K. and Bono, H. (2023) ‘Danger analysis: Risk-averse on/off-target assessment for CRISPR editing without a reference genome’, Bioinformatics Advances, 3(1). doi:10.1093/bioadv/vbad114.