Reviewed by Danielle Ellis, B.Sc.Nov 7 2023
In 2015, the first instances of tar spot were reported, marking the emergence of a devastating disease in corn. This ailment swiftly spread throughout the United States and Canada, resulting in substantial yield losses amounting to an estimated $1.2 billion in 2021 alone.
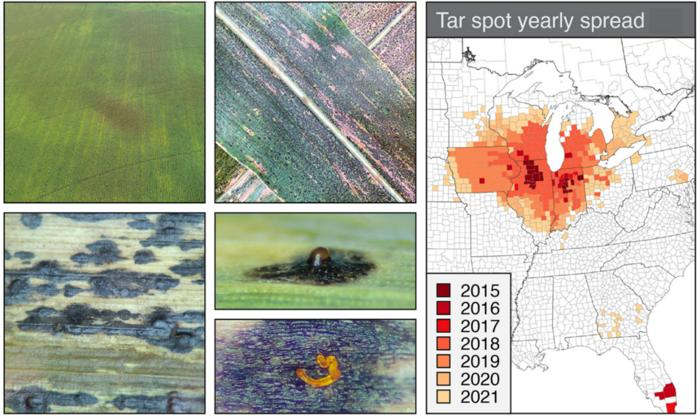 Tar spot symptoms (left panel) - Aerial view of infected tar spot field (top left). Close up of infected leaf (top right). Tar spot lesions (bottom left). Sporulating lesion (bottom right). Tar spot yearly spread from 2015 to 2021 (right panel). Image Credit: Joshua S. MacCready et al.
Tar spot symptoms (left panel) - Aerial view of infected tar spot field (top left). Close up of infected leaf (top right). Tar spot lesions (bottom left). Sporulating lesion (bottom right). Tar spot yearly spread from 2015 to 2021 (right panel). Image Credit: Joshua S. MacCready et al.
The name “tar spot” originates from its distinctive symptoms, which resemble dark, tar-like splotches on corn leaves. However, these marks are actually brown lesions caused by the fungal pathogen Phyllachora maydis.
This destructive pathogen presents a challenge for researchers because it cannot survive outside of its corn host. Consequently, human knowledge about the mechanisms driving its disease cycle, such as spore formation, reproduction, and plant infection, remains limited.
To bridge this knowledge gap, a recent study led by Joshua MacCready and Emily Roggenkamp, affiliated with Martin I. Chilvers’s laboratory at Michigan State University, successfully generated a high-quality genome for P. maydis. Additionally, this study marked the first comprehensive examination of global gene expression during a tar spot disease infection.

Image Credit: Dejan82/Shutterstock.com
The study, published in the journal Molecular Plant-Microbe Interactions, uncovered a crucial revelation: the fungus undergoes sexual reproduction early in the formation of the lesions, a process necessitating two individuals of different mating types.
This discovery holds significance because sexual reproduction equips the fungus with increased genetic variability, potentially leading to the development of more virulent or fungicide-resistant strains.
Similar to many other fungal pathogens, P. maydis relies on the secretion of proteins, referred to as effectors, to manipulate and counteract plant immune responses. This study identified over 100 effectors, many of which are unique to this fungal pathogen and are highly expressed during infection.
This suggests that these proteins likely play a novel role during infection, warranting further investigation to elucidate their specific functions.
The newly established genome and the insights gained through gene expression analysis present a valuable resource for fellow researchers studying tar spot disease in corn.
This study lays the groundwork for future studies to examine effectors, pathogenesis, and the lifecycle of this pathogen.”
Martin I. Chilvers, Michigan State University
Equipped with a deeper comprehension of this fungus, these researchers aspire to deploy enhanced strategies for managing this influential pathogen.
Source:
Journal reference:
MacCready, J. S., et al. (2023) Elucidating the Obligate Nature and Biological Capacity of an Invasive Fungal Corn Pathogen. Molecular Plant-Microbe Interactions. doi.org/10.1094/MPMI-10-22-0213-R