Reviewed by Danielle Ellis, B.Sc.Nov 28 2023
A recent investigation led by Professor Sigal Ben Yehuda and her team at Hebrew University has brought to light a fascinating aspect of bacterial dormancy.
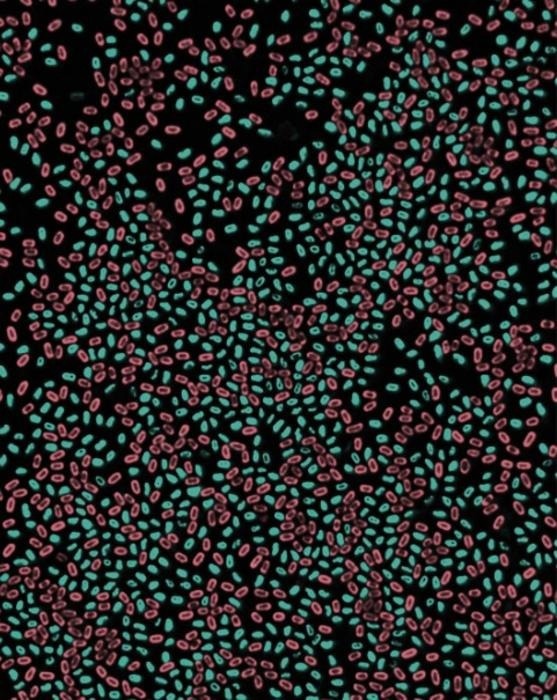 Fluorescence microscopy images of Bacillus subtilis spores harboring GFP or Scarlet fusion to a DNA packaging protein (SspA). Image Credit: Bing Zhou.
Fluorescence microscopy images of Bacillus subtilis spores harboring GFP or Scarlet fusion to a DNA packaging protein (SspA). Image Credit: Bing Zhou.
The study elucidates the mechanism by which dormant bacterial spores maintain and activate a persistent transcriptional program upon revival, revealing an extraordinary genetic memory system.
This revelation is crucial as it unveils the mechanisms governing the retention of vital genetic information by these organisms during prolonged periods of dormancy. Understanding this process not only provides insights into bacterial survival under harsh conditions but also carries broader implications.
The findings could offer an understanding of sustaining long-term transcriptional programs in various organisms, potentially influencing fields such as microbiology, biotechnology, and medicine. This knowledge may pave the way for strategies to control pathogens, improve biotechnological processes, and deepen the comprehension of dormant states across different life forms.
Spores, robust and protective structures formed by certain microorganisms like bacteria and fungi, act as a survival mechanism against adverse conditions. Bacterial spores, known to be among the longest-living cellular forms on Earth, can revive even after millions of years of quiescence.
Image Credit: Kateryna Kon/Shutterstock.com
These spores encapsulate the organism’s genetic material and essential components, remaining dormant until conditions become favorable for germination.
Understanding spores is crucial in various fields, providing insights into survival strategies and potential applications in microbiology, agriculture, and biotechnology.
Published in Molecular Cell, the study underscores the discovery of a central chromosomal domain within dormant spores. This domain houses core RNA polymerase (RNAP), which remains bound to specific intergenic promoter regions during dormancy. These regions exert control over genes essential for crucial cellular functions, including the production of rRNAs and tRNAs.
Upon emergence from dormancy, the RNA polymerase inside these spores promptly initiates the replication of vital genetic instructions. It recruits necessary components for transcription, such as sigma factors, swiftly activating essential genes necessary for cellular functions.
The study also identified a similar process in disease-causing bacteria that form spores, suggesting a common strategy among various organisms to reinitiate functions after dormancy.
Additionally, the research unveiled the pivotal role of spore DNA-compacting proteins in this process. Mutants lacking these proteins exhibited scattered RNAP localization, leading to disorganized gene expression during germination. This underscores the significance of maintaining proper chromosomal structure in preserving the transcriptional program crucial for spore revival.
Our research suggests that the structure of the spore chromosome is designed to uphold a blueprint for gene activity by pausing RNA polymerase, in a standby mode, ready to resume gene expression when conditions favor revival. This mechanisms relevance might extend beyond bacteria, offering valuable insights into maintaining enduring gene activity plans across various organisms that undergo dormant life stages.”
Sigal Ben Yehuda, Professor, Hebrew University
This study marks a significant advance in understanding the complex mechanisms underlying bacterial dormancy and resurgence. Its ramifications extend across a range of fields, encompassing microbiology and offering potential applications in biotechnology and medicine.
Source:
Journal reference:
Zhou, B., et al. (2023) Dormant bacterial spores encrypt a long-lasting transcriptional program to be executed during revival. Molecular Cell. doi.org/10.1016/j.molcel.2023.10.010.