Reviewed by Danielle Ellis, B.Sc.Nov 30 2023
Spatial transcriptomics (ST) has emerged as a crucial technique for mapping gene expression across tissue sections while preserving vital locational data, setting it apart from traditional methods like bulk or single-cell RNA sequencing.
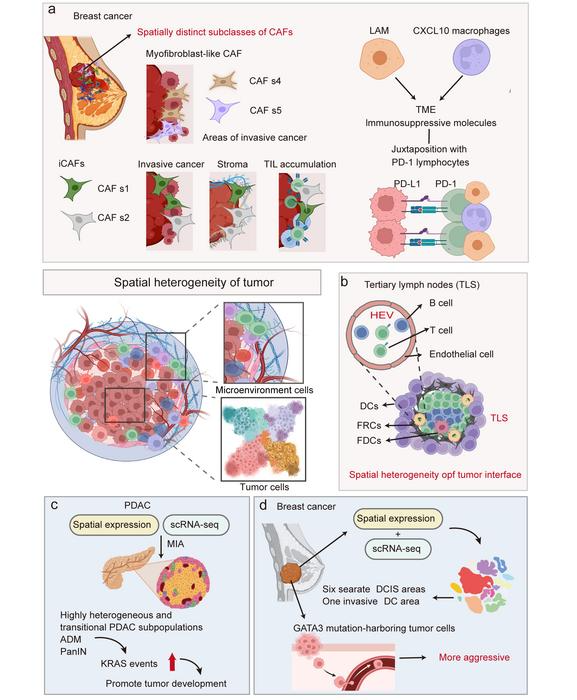 Spatial transcriptomics techniques facilitate the study of tumor microenvironment heterogeneity and tumor heterogeneity. a ST examines the diversity within cancer-associated fibroblasts and immunosuppressive molecules within the microenvironment of breast cancer. b ST enables the identification and comprehensive exploration of unique tumor microenvironment regions, such as the tumor interface and tertiary lymph nodes. c ST analyzes the spatial distribution of PDAC-associated heterogeneity, identifying highly heterogeneous and transitional PDAC subpopulations. d ST applied to breast cancer biopsy identified that tumor cells harboring GATA3 mutations became more invasive, revealing the spatial heterogeneity of breast cancer. Image Credit: Biorender.com for image resources
Spatial transcriptomics techniques facilitate the study of tumor microenvironment heterogeneity and tumor heterogeneity. a ST examines the diversity within cancer-associated fibroblasts and immunosuppressive molecules within the microenvironment of breast cancer. b ST enables the identification and comprehensive exploration of unique tumor microenvironment regions, such as the tumor interface and tertiary lymph nodes. c ST analyzes the spatial distribution of PDAC-associated heterogeneity, identifying highly heterogeneous and transitional PDAC subpopulations. d ST applied to breast cancer biopsy identified that tumor cells harboring GATA3 mutations became more invasive, revealing the spatial heterogeneity of breast cancer. Image Credit: Biorender.com for image resources
Unlike these approaches, ST simultaneously captures gene expression profiles and the spatial coordinates of cells, unveiling hidden aspects of cellular heterogeneity, organizational patterns, and molecular interactions crucial to tissue dynamics.
Furthermore, ST can seamlessly integrate with other “omics” strategies, providing a comprehensive perspective on biological entities at various levels of detail. Over the course of its existence, ST has introduced novel methods that enhance throughput and resolution, promising significant progress in understanding biological complexities.
This review systematically categorizes and compares various ST techniques based on their foundational principles and procedural intricacies, offering a comprehensive assessment of the diverse approaches within the field.
The evaluation underscores the inherent trade-off between acquiring high-resolution data and maintaining a high-throughput workflow. Recent innovations, like Stereo-seq, show considerable strides in resolving this trade-off, yet the pursuit of the gold standard—precise localization of each transcript at the cellular level—persists.
Image Credit: crystal light/Shutterstock.com
Moreover, the review highlights the utility of ST across various biomedical research fields, including developmental biology, neuroscience, immunology, and oncology.
ST has played a pivotal role in unraveling intricate cellular dynamics within tissue architectures, shedding light on mutual interactions and their crucial functions in complex biological processes, thereby providing transformative insights into conditions ranging from cancers to neurodegenerative diseases.
The examination extends to ST's proficiency in deciphering the complexities of the tumor microenvironment, addressing persistent challenges such as intratumoral heterogeneity that hinder effective oncological interventions.
Additionally, the review discusses current limitations, such as the diverse methodologies leading to varied file formats and data structures, making data and protocol sharing challenging. Harmonizing spatially resolved data across intra-omics and cross-omics layers poses a significant hurdle.
Finally, the review explores prospective trajectories for advancing spatial transcriptomics, emphasizing the need to enhance spatial resolution, expand gene coverage, and simplify operational complexities.
There is an urgent call for the development of new computational tools to improve processing and integration. Summarizing recent advancements in historical, technical, and application contexts, this review serves as a valuable reference for researchers, guiding them in selecting spatial transcriptomics methodologies tailored to their unique biological inquiries and experimental conditions.
Source:
Journal reference:
Zhou, R., et al. (2023) Spatial transcriptomics in development and disease. Molecular Biomedicine. doi.org/10.1186/s43556-023-00144-0.