Reviewed by Danielle Ellis, B.Sc.Jan 2 2024
Professor Wen-Bin Zhang of the College of Chemistry and Molecular Engineering at Peking University and the Beijing Academy of Artificial Intelligence, along with Dr. Jing Fang of the same institution, are leading the study. Two mechanically linked polypeptide rings that fold cooperatively into a compact and integrated structure—an incredibly uncommon occurrence in nature—are referred to as single-domain protein catenane.
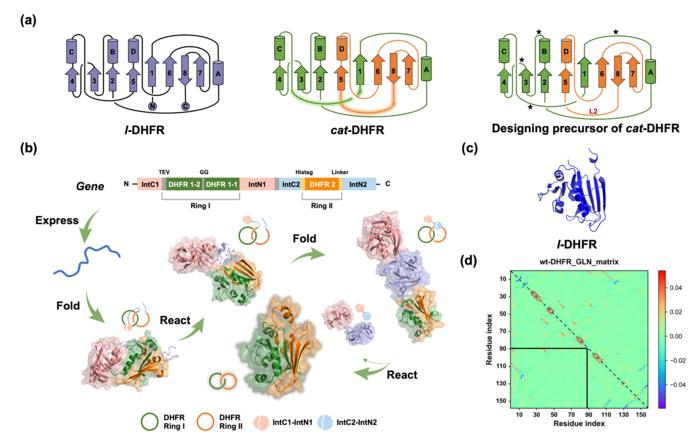 (a) Protein topological diagram of l-DHFR (left) and cat-DHFR (middle), and the retrosynthetic analysis of cat-DHFR (right). Numbers 1–8 and letters A–D represent the β-sheet and α-helix, respectively, from N- to C- termini in consecutive order. The split site (residues 88 and 89) is located at the loop region between α-helix-C and β-sheet-5. The highlighted lines are the linkers generated when forming the cat-DHFR. The star denotes the possible position for ring I closure (at the same and opposite sides) and split-intein insertion. L2 is the linker newly introduced to ring II of cat-DHFR. (b) Scheme of the cat-DHFR biosynthesis process using programmed post-translation processing events. DHFR1 is circularly permutated, and the corresponding sequences are denoted by DHFR1-1 and DHFR1-2. The TEV recognition site and a GG linker were inserted into ring I. The His-tag and a variable linker (together, they are L2) were inserted into ring II. (c) Structure prediction (https://robetta.bakerlab.org/) of l-DHFR. (d) The Gaussian linking number (GLN) matrix of wt-DHFR. It comprises GLN values between all neighboring residue pairs within the same chain. The sum of all the cells within the boxed sub-matrix corresponds to the GLN value between the two subchains, which provides a quantitative metric of the extent of their entanglement. Image Credit: Science China Press
(a) Protein topological diagram of l-DHFR (left) and cat-DHFR (middle), and the retrosynthetic analysis of cat-DHFR (right). Numbers 1–8 and letters A–D represent the β-sheet and α-helix, respectively, from N- to C- termini in consecutive order. The split site (residues 88 and 89) is located at the loop region between α-helix-C and β-sheet-5. The highlighted lines are the linkers generated when forming the cat-DHFR. The star denotes the possible position for ring I closure (at the same and opposite sides) and split-intein insertion. L2 is the linker newly introduced to ring II of cat-DHFR. (b) Scheme of the cat-DHFR biosynthesis process using programmed post-translation processing events. DHFR1 is circularly permutated, and the corresponding sequences are denoted by DHFR1-1 and DHFR1-2. The TEV recognition site and a GG linker were inserted into ring I. The His-tag and a variable linker (together, they are L2) were inserted into ring II. (c) Structure prediction (https://robetta.bakerlab.org/) of l-DHFR. (d) The Gaussian linking number (GLN) matrix of wt-DHFR. It comprises GLN values between all neighboring residue pairs within the same chain. The sum of all the cells within the boxed sub-matrix corresponds to the GLN value between the two subchains, which provides a quantitative metric of the extent of their entanglement. Image Credit: Science China Press
Artificial entanglement was introduced into this design by rewiring the connection between secondary motifs. Synthesis was easily completed by a sequence of simplified, planned post-translational processing events in cells, without the need for any extra in vitro processes.
A detailed characterization of the single-domain catenane cat-DHFR was performed. Its topology was clearly demonstrated by data from combined SDS-PAGE, SEC, LC-MS, IMS-MS, and proteolytic digestion investigations. With a Tm that is 6 °C higher than the linear control, the cat-DHFR has improved anti-aggregation characteristics.
Better heat resistance than l-DHFR is exhibited by cat-DHFR, despite its lower affinity for the substrate and cofactor leading to a reduction in catalytic activity. Cat-DHFR maintained more than 70% of its catalytic activity even after 10 minutes of incubation at 70 °C, while the linear control almost lost all of its activity.
Image Credit: Christoph Burgstedt/Shutterstock.com
The researchers believe that this approach could be broadly applicable to other single-domain proteins, such as those that have folds that are entirely different from or comparable to DHFR. The availability of these single-domain protein catenanes makes it easier to understand how topology affects links between structure and property.
The findings also suggest that it is feasible to transfer the existing linear protein universe onto single-domain protein catenanes that have additional advantages and well-preserved functionalities, hence providing new opportunities for protein molecules. These topological proteins are multi-chain, multi-dimensional structures that go beyond the linear paradigm of natural protein molecules.
They have vast design possibilities, good evolvability, and the functional advantages of topology. They have enormous promise as a novel class of protein molecules with a wide range of uses, such as industrial enzymes, antibodies, cytokines, and biomaterials.
Source:
Journal reference:
Fang, J., et al. (2023). A Single-domain Protein Catenane of Dihydrofolate Reductase. National Science Review. doi.org/10.1093/nsr/nwad304