The last ten years have seen significant advancements in CRISPR/Cas systems. These precise genome editing tools can be used for more than just gene therapy and transgenic crop development.
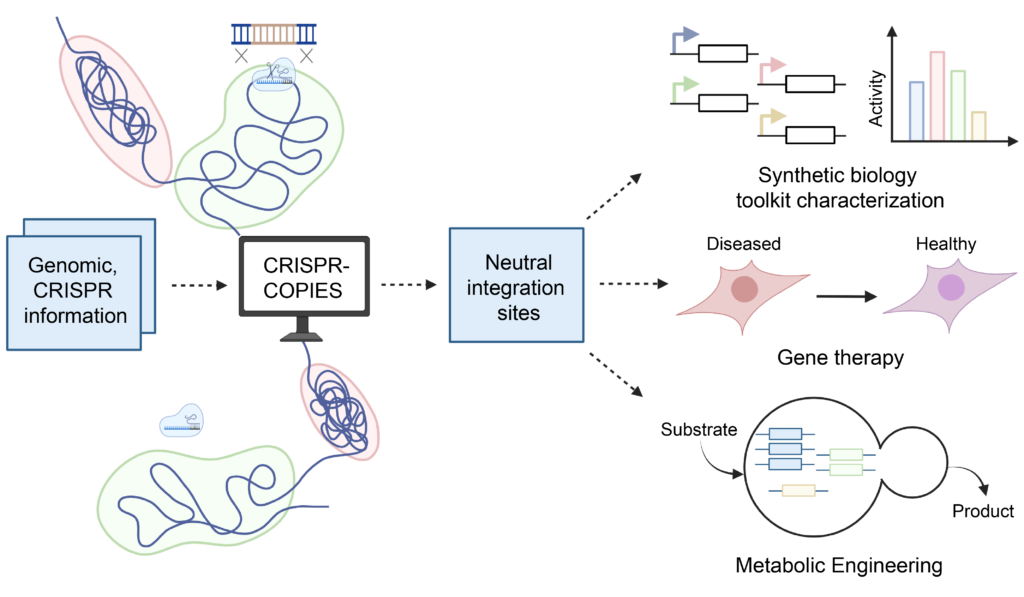
Image Credit: CABBI
Furthermore, scientists at the Center for Advanced Bioenergy and Bioproducts Innovation (CABBI) are expanding the capabilities and usability of CRISPR with the creation of CRISPR-COPIES.
CRISPR-COPIES is a tool that can quickly identify appropriate chromosomal integration sites for genetic engineering in any organism. It will accelerate our work in the metabolic engineering of non-model yeasts for cost-effective production of chemicals and biofuels.”
Huimin Zhao, Steven L. Miller Chair of Chemical and Biomolecular Engineering, University of Illinois
The ability of scientists to comprehend and work with genetic information has been transformed by gene editing. Through this type of genetic engineering, scientists can give an organism new characteristics like pest resistance or the capacity to produce a useful biochemical.
Researchers can make precise, targeted genetic edits using CRISPR/Cas systems. However, finding the best integration sites for these edits in the genome has proven to be a crucial and largely unresolved issue.
In the past, researchers would usually manually screen for possible integration sites to determine where to target their edits. After that, they would test the site by integrating a reporter gene in order to evaluate its levels of gene expression and cellular fitness. It takes a lot of time and resources to complete.
CRISPR-COPIES, a Computational Pipeline for identifying CRISPR/Cas-facilitated intEgration Sites, was created by the CABBI team in response to this difficulty. In two to three minutes, this tool can identify genome-wide neutral integration sites for most bacterial and fungal genomes.
Finding the integration site in the genome manually is like searching for a needle in a haystack. However, with CRISPR-COPIES, we transform the haystack into a searchable space, empowering researchers to efficiently locate all the needles that align with their specific criteria.”
Aashutosh Boob, Study Primary Author and Student, University of Illinois
The researchers used HEK 293T cells, Saccharomyces cerevisiae, and Cupriavidus necator to characterize integration sites to illustrate the adaptability and scalability of CRISPR-COPIES in their paper published in Nucleic Acids Research.
They engineered cells to produce more 5-aminolevulinic acid, a useful biochemical used in agriculture and the food industry, using integration sites discovered by CRISPR-COPIES.
Furthermore, the group has developed an intuitive web interface for CRISPR-COPIES. Researchers with little to no experience with bioinformatics can still use this incredibly user-friendly application.
The engineering of non-model yeasts to generate chemicals and fuels from plant biomass is one of CABBI’s main goals. It is difficult to economically produce biofuels and bioproducts from inexpensive feedstocks on a large scale, though, because conventional genome-editing techniques are laborious and lack genetic tools.
CRISPR-COPIES offers a simplified pipeline that makes identifying stable integration sites throughout the genome easier by allowing researchers to identify genomic loci for targeted gene integration quickly. Additionally, it does away with the need for manual labor when designing parts for DNA integration mediated by CRISPR/Cas.
The tool can be used in crop engineering to boost environmental resilience, pest resistance, and/or biomass yields. Using the yeast S. cerevisiae, for example, CRISPR-COPIES can be used to engineer cells with much higher yields for biomass conversion to useful chemicals.
This flexible software helps researchers save time and money by streamlining and expediting the strain construction process. Its application in strain engineering for biochemical production and transgenic crop development can benefit researchers globally, in both academic and industrial contexts.
Co-authors of this work include Manan Jain and Vassily Petrov, software developers at the Carl R. Woese Institute for Genomic Biology (IGB); Stephan Lane, biofoundry manager at the IGB; CABBI postdoc Shih-I Tan; and Ph.D. candidates in bioengineering and ChBE, as well as visiting students Pattarawan Intasian and Zhixin Zhu.
Source:
Journal reference:
Boob, G. A., et.al. (2024) CRISPR-COPIES: an in silico platform for discovery of neutral integration sites for CRISPR/Cas-facilitated gene integration. Nucleic Acids Research. doi.org/10.1093/nar/gkae062