Plant cell analysis has been completely transformed by recent advances in single-cell RNA sequencing (scRNA), such as the recently created “RevGel-seq” technique. This method solves problems with protoplast isolation and streamlines procedures without the need for specialized equipment.
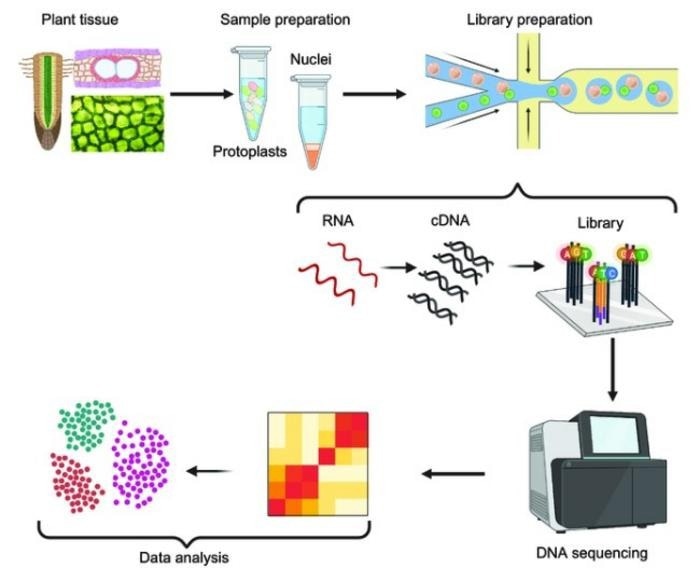 Starting from whole plant tissues and dissociating cells (protoplasts) or nuclei from the tissue, RNA extraction, cDNA synthesis by reverse transcription, library preparation followed by sequencing, expression abundance estimation, and cell-type identification. Redrawn from Yip S. H., et al. Image Credit: BioDesign Research.
Starting from whole plant tissues and dissociating cells (protoplasts) or nuclei from the tissue, RNA extraction, cDNA synthesis by reverse transcription, library preparation followed by sequencing, expression abundance estimation, and cell-type identification. Redrawn from Yip S. H., et al. Image Credit: BioDesign Research.
To facilitate the proper selection of scRNA methods for various plant samples, a multinational team of researchers is now reviewing this and other recent developments in plant scRNA sequencing.
Understanding the nuances of individual plant cells has proven to be a difficult task in the field of plant biology, in part because of the peculiar structure of these cells, which are encased in stiff cell walls. This difficulty has consequently made it more difficult to isolate intact nuclei or protoplasts, which are necessary for a thorough examination.
Now, single-cell RNA (scRNA) sequencing has made revolutionary strides in understanding and analysis of plant biosystems possible, even though traditional RNA sequencing has been crucial in helping researchers comprehend the complexities of plant cells.
Furthermore, the capacity to overcome analytic challenges unique to plants has been strengthened by additional technological developments in scRNA sequencing, such as the creation of protocols for cell isolation, library preparation, and sequencing technologies.
Studies on the usefulness and practical applicability of scRNA sequencing have been reviewed in the past, but few have looked at the technological advancements related to scRNA sequencing.
However, a thorough examination of the most recent developments and paradigm shifts in plant systems biology has now been conducted by researchers and published in the journal BioDesign Research.
Due to their intricate cellular architecture, plant systems require novel approaches to reveal the mysteries that reside within their cells. The review discusses the difficulties and prospective opportunities for the future while also capturing these recent advancements in scRNA technology.
The review first goes over the widely accepted techniques for single-cell transcriptomics, which involve both computational and experimental elements. It also goes into detail about the overall scRNA sequencing workflow.
The use of a microfluidics platform provides a primary workflow for single-cell isolation, separation, and analysis in light of the difficulties and constraints associated with protoplast preparation and nucleus isolation techniques.
Additionally, the review highlights that while the research objective determines which of the protoplast and nuclei to use for plant scRNA sequencing, researchers should carefully consider a number of factors, including the research goal, plant species, and other trade-offs, when determining the most appropriate method.
The review then discusses the developments in scRNA sequencing library construction and sequencing, emphasizing notable successes like Illumina sequencing technologies, the Nextera XT DNA Library Preparation Kit, and the BD Rhapsody system.
The researchers then review the various stages of scRNA sequencing before discussing the most current advancements in scRNA databases.
For example, PlantscRNAdb, which includes 26,326 marker genes, was created to analyze scRNA-seq data in plants. In contrast, the Plant Cell Marker DataBase includes 81,117 cell marker genes from 263 cell types in 22 tissues from six different plant species.
The advancements made by scientists to overcome the shortcomings with plant scRNA databases, like “scPlantDB,” an extensive database made by He, et al. covering around 2.5 million cells across 17 plant species.
The researchers continue by going into detail about the uses of scRNA sequencing in plant systems biology and how it has been applied to study plant biology at the cellular level.
This has allowed researchers to learn more about the molecular mechanisms underlying plant development, responses of plants to biotic and abiotic stresses, plant epigenetic regulation, determination of cell fate, and organogenesis, among other topics.
The researchers highlight a recent breakthrough development that aims to optimize protoplast isolation methods and establish standardized single-cell RNA sequencing (scRNA-seq) processes across various laboratories, while also discussing the challenges and opportunities for further development of scRNA sequencing.
Using a multimodal strategy, the research aimed to address the complex world of plant cells by fusing cutting-edge scRNA-seq technologies with conventional techniques. Sample preparation became an exacting procedure that paid close attention to factors like temperature, osmotic potential, and the length of the enzyme treatment.
The study was noteworthy for its exploration of novel scRNA-seq technologies, such as RevGel-seq, which was initially developed for animal systems and is now being applied to plant research.
RevGel-seq, a recently developed breakthrough in scRNA-seq methods, is truly a game-changer. Unlike traditional methods that rely on specific single-cell RNA instruments during sample preparation, RevGel-seq operates on cell-barcoded bead complexes. This innovation sample preparation for scRNA-seq in human and mouse cell not only convenient but also highly efficient.”
Dr. Xiaohan Yang, Study Lead Researcher, Oak Ridge National Laboratory
Additionally, RevGel-seq removes the requirement for particular instruments, allowing for flexibility in sample collection and processing at various times or locations. Without a doubt, the outcomes exceeded predictions, demonstrating the potential of RevGel-seq to transform the way single-cell RNA sequencing is carried out in plant research.
The recent developments covered in this review will have a big impact on plant biology going forward. Even though scRNA sequencing has become a ground-breaking technique in the fields of synthetic biology and plant systems biology, the review clarifies the way forward for resolving current issues and provides guidance for upcoming investigations.
It also emphasizes how important it is to combine scRNA sequencing technologies with computational tools and other omics technologies to improve the comprehension of intricate cellular interactions in plants.
In summary, the review points to a promising future for plant biology, with single-cell RNA sequencing for plant cells providing new insights into their internal mechanisms. It can help researchers make significant advancements toward a more sustainable future in addition to addressing present obstacles.
Source:
Journal reference:
Islam, T., M. D., et al. (2024) Advances in the application of single-cell transcriptomics in plant systems and synthetic biology. Biodesign Research. doi.org/10.34133/bdr.0029