Drug developers have long faced challenges in reaching targets embedded within the cell membrane because of the membrane's complex biochemical properties. Chemists at Scripps Research have now shown that novel, specially-made proteins are capable of effectively reaching these “intramembrane” targets.
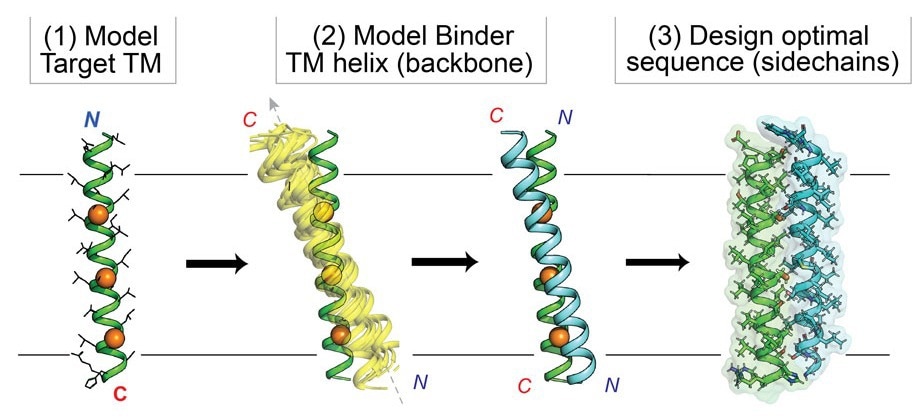 The computational process, or “workflow,” for the custom design of synthetic proteins aimed at intramembrane targets. Image Credit: Marco Mravic, Scripps Research.
The computational process, or “workflow,” for the custom design of synthetic proteins aimed at intramembrane targets. Image Credit: Marco Mravic, Scripps Research.
The researchers used a novel computer-based method to design novel proteins that target the membrane-spanning region of the erythropoietin (EPO) receptor, which regulates red blood cell production and can malfunction in cancers. The study was published in the journal Nature Chemical Biology.
Apart from the newly discovered EPO-targeting compounds, the research produced a general computational procedure, or “workflow,” that facilitates the efficient custom design of proteins with intramembrane targets.
Now, the researchers are applying their methodology to create novel intramembrane-targeting therapies for various diseases.
This work opens up many new possibilities for the modulation of targets within the cell membrane, including for therapeutic applications and understanding signaling mechanisms across cell biology.”
Marco Mravic, Ph.D., Assistant Professor and Study Co-Corresponding Author, Department of Integrative Structural and Computational Biology, Scripps Research
Other Co-Corresponding Authors include Daniel DiMaio, MD, Ph.D., of the Yale School of Medicine and William DeGrado, Ph.D., of the University of California, San Francisco School of Pharmacy, where Mravic was previously a Ph.D. student.
A major goal of synthetic biology is to design proteins with biological activity here, we report the design and testing of a small protein that specifically perturbs the activity of a much larger protein receptor involved in blood cell growth and differentiation.”
Daniel DiMaio, Professor, Department of Genetics and Molecular Biophysics, Yale School of Medicine
DiMaio said, “We accomplished this by targeting the segment of the receptor that crosses the cell membrane. Because many cell proteins contain structurally conserved membrane-spanning segments, this general approach may be applicable to many other protein targets and provides a new tool to modulate the behavior of cells.”
Reaching intramembrane targets has long been a biomedical goal because many proteins, particularly receptor proteins, have membrane-bound functional domains. These proteins play important roles in nearly every aspect of health and illness.
Intramembrane targets, however, are not typical targets. Generally speaking, cell membranes consist of two layers of closely spaced “lipid” molecules related to fat that have various complex and unique properties, including the ability to repel water.
Compared to the wet areas of cell surfaces or interiors, this makes the intramembrane space a far more difficult target for drug designers.
Mravic said, “There have been very few successful examples of drugs that target this space inside the membrane.”
Those few successes, which include treatments for cystic fibrosis and low-blood-platelet disorders, have resulted from close mimicry of proteins known to interact with partner proteins within the cell membrane or from blind screenings of large compound libraries.
On the other hand, Mravic and associates aimed to create entirely new proteins, referred to as peptides, that would target intramembrane protein targets in a variety of fresh and inventive ways.
To achieve that, they had to push the boundaries of computational techniques by fusing conventional physics-based predictions of protein interactions with "data mining" of known protein-to-protein interactions in membranes.
In the end, Mravic and his associates created the first proteins that bind the membrane-spanning region of the EPO receptor in a novel manner not found in nature. Unlike previous methods, the team demonstrated that these proteins very specifically and potently block the function of the receptor.
Researchers looking for novel approaches to inhibit the EPO receptor, which tumor cells frequently aberrantly activate to sustain their growth and survival, may be most interested in the results immediately. However, the study primarily serves as proof of principle for a novel and adaptable intramembrane targeting strategy, according to Mravic and his associates.
Mravic said, “We intend to use this approach to target membrane proteins across multiple biological processes and disease areas, including cancers, immune disorders, and pain.”
Additionally, he anticipates that the computational workflow he created for the project will serve as a general membrane-targeting drug design accelerator.
Before, the process basically involved two people in a dark room looking at a computer screen and saying, Yeah, I think this looks better than that, and now, we have automated a lot of that molecule design process and decision making in the computer. Being more modular, flexible, and streamlined allows the method to be more accessible for a broader range of scientists.”
Marco Mravic, Ph.D., Assistant Professor and Study Co-Corresponding Author, Department of Integrative Structural and Computational Biology, Scripps Research
Source:
Journal reference:
Mravic, M., et al. (2024) De novo-designed transmembrane proteins bind and regulate a cytokine receptor. Nature Chemical Biology. doi.org/10.1038/s41589-024-01562-z