Researchers at EPFL have opened up a world of possibilities in the computational design of molecular interactions for biomedicine by using deep learning to create new proteins that bind to complexes involving other small molecules, such as hormones or medications.
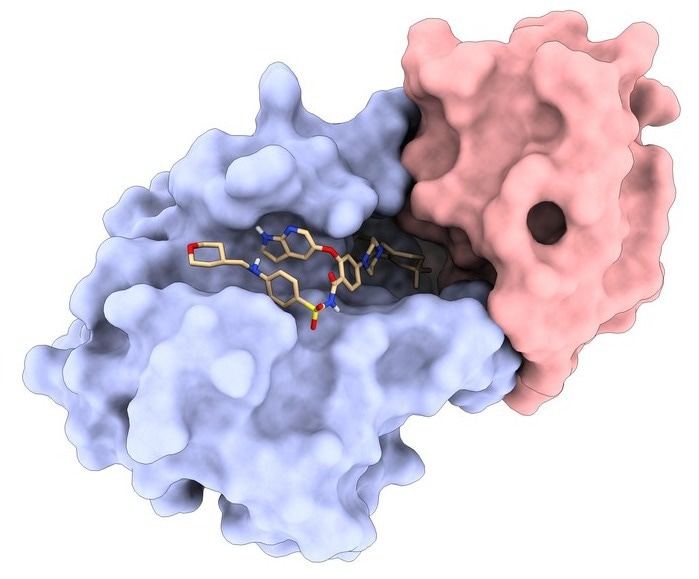 One of the LPDI's de novo protein binders (red) bound to the protein Bcl2 (blue) in complex with FDA-approved drug Venetoclax (beige). Image Credit: LPDI EPFL
One of the LPDI's de novo protein binders (red) bound to the protein Bcl2 (blue) in complex with FDA-approved drug Venetoclax (beige). Image Credit: LPDI EPFL
In 2023, researchers from the School of Engineering and the School of Life Sciences’ Laboratory of Protein Design and Immunoengineering (LPDI) at EPFL, led by Bruno Correia, published a deep-learning pipeline in Nature. This innovative approach enables the creation of new proteins designed to interact with therapeutic targets.
By leveraging the capabilities of MaSIF, a machine-learning framework, scientists can rapidly analyze millions of proteins to identify optimal matches based on their chemical and geometric surface properties. This advancement facilitates the design of protein-protein interactions crucial for cellular regulation and therapeutic applications.
A year and a half later, the team reported a significant breakthrough, also published in Nature. They successfully used MaSIF to design proteins that bind to complexes of proteins and small molecules, such as hormones or drugs. These small molecule-protein complexes can act as precise "on" or "off" switches, modulating cellular processes like DNA transcription and protein degradation by altering their surface characteristics—termed "neosurfaces."
“Our idea was to engineer an interaction where a small molecule brings two proteins together. While some approaches focus on screening for such small molecules, we aimed to design a novel protein to bind to a specific protein-drug complex.”
– Anthony Marchand, Scientist and Co-First Author, LPDI, EPFL
Remarkably, MaSIF demonstrated its ability to generalize surface "fingerprints" trained on proteins to the neosurfaces formed by protein-drug complexes. This sensitivity and adaptability to small molecules set it apart from most learning-based protein design systems, which typically rely only on natural amino acid building blocks.
Small but Powerful Insights
Protein binding might seem as simple as fitting puzzle pieces together, but surface variations make predicting binding sites and events highly challenging. As in their earlier work, the team used MaSIF to generate "fingerprints" of surface features like hydrophobicity, shape, and charge. These fingerprints were then used to digitally graft protein fragments onto scaffolds, select the best-fitting binders, and identify complementary surfaces in a database.
“We assumed that when a small molecule binds to a protein, it creates a neosurface by altering the protein’s features. MaSIF captured this change with remarkable sensitivity,” explained Arne Schneuing, PhD student and co-author at LPDI.
To validate their approach, the researchers designed protein binders for three drug-bound complexes involving progesterone, the leukemia drug Venetoclax, and the antibiotic Actinonin. The MaSIF-designed binders exhibited high affinity and accurately recognized these complexes. This success stemmed from MaSIF’s ability to map small molecule features onto the same descriptor space it uses for proteins, demonstrating its versatility.
“MaSIF uses about 70,000 parameters—significantly fewer than the billions used in larger deep-learning systems like ChatGPT. By focusing only on key surface features, we achieve a high level of abstraction, providing the system just the essentials to solve the problem,” added Schneuing.
Applications in CAR-T Therapy
One promising application of this research is in improving the precision of cell-based therapies like chimeric antigen receptor (CAR-T) therapy, where a patient’s T cells are engineered to better target cancer. Despite its potential, CAR-T therapy can lead to unintended side effects if engineered cells attack off-target sites or lose their effectiveness.
In a proof-of-concept experiment, the EPFL team demonstrated that a Venetoclax-inducible system designed with MaSIF successfully activated CAR-T cells to kill tumor cells in vitro.
“If we can precisely control the timing and location of cell-based therapies using small molecule switches, we can significantly enhance their safety and effectiveness,” concluded Stephen Buckley, LPDI PhD student and co-first author.
Source:
Journal reference:
Marchand, A., et al. (2025) Targeting protein–ligand neosurfaces with a generalizable deep learning tool. Nature. doi.org/10.1038/s41586-024-08435-4