The concept of antisense oligonucleotide technologies is based on blocking mRNA activity, leading to specific inhibition of undesirable gene expression.
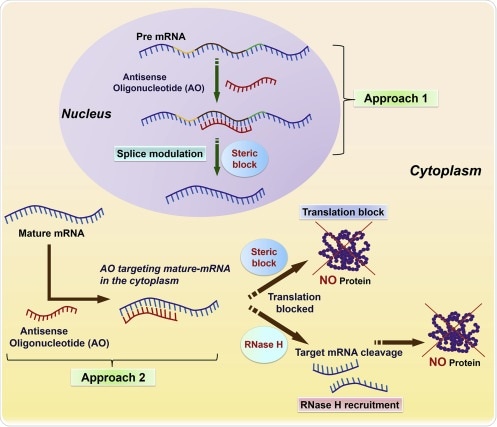
Image Credit: https://www.cell.com/molecular-therapy-family/nucleic-acids/fulltext/S2162-2531(18)30301-9
For a long time, this is considered to be an ideal strategy to use new genomic knowledge for drug discovery and development. In recent years, antisense oligonucleotide technologies have been widely used as potent and promising tools for the inhibition of gene expression in a sequence-specific way.
Molecular mechanisms of antisense oligonucleotides
Antisense oligonucleotides are short sequences of nucleic acid, which are consisted of 7 – 30 nucleotides. They can bind to a specific region of a target mRNA according to Watson–Crick base pairing rules, inhibiting unwanted gene expression.
The mechanism of action by which antisense oligonucleotides inhibit mRNA translation is based on the chemistry of oligonucleotides. For instance, oligonucleotides consisting of regions of DNA such as phosphorothioate phosphodiester and chimeric work through recruiting endogenous ribonuclease H (RNase H), which is a ubiquitously expressed ribonuclease that preferentially binds to RNA–DNA duplexes and initiates cleavage of the RNA strand.
Modified antisense oligomers, which do not use ribonuclease H such as peptide nucleic acids, morpholino, have the ability to inhibit protein synthesis via hybridizing in the 5′ UTR or near the AUG start codon, which can block assembly of the ribosome. They also may inhibit splicing when hybridized to intron-exon junctions.
The mechanisms of action of antisense oligonucleotides must be demonstrated by direct inhibition of the target gene at the RNA or protein level by several antisense oligonucleotides directed against different sites on the target RNA and by lack of inhibition by several scrambled and mismatch control oligonucleotides.
The development in antisense oligonucleotide chemistry and methodologies for the delivery of oligonucleotides to cells has led to reproducible results demonstrating that antisense oligonucleotides have strong and specific inhibition of gene expression. Antisense technology is currently considered to be a proven technology for an explanation of target-gene function and the recognition of novel drug targets
The future of antisense oligonucleotide technologies
In the previous decade, specifically in 1996, there was only a handful of antisense molecules used in clinical trials. Out of these six trials, five were only in Phase I. Yet, in recent years, there was tremendous growth in the number of antisense related clinical trials.
Food and Drug Administration (FDA) has approved one antisense oligonucleotide molecule, for local administration to treat cytomegalovirus (CMV) retinitis, demonstrating that antisense drugs could be used effectively in the treatment of local disease. To date, there are more than fifty antisense oligonucleotides in trials for various diseases.
Most of these drugs are oligonucleotides formulated to prevent gene expression via an RNase H mechanism. Moreover, more than thirty biotechnology and pharmaceutical companies have announced an interest in, or have a running drug development program in RNAi-based therapeutics.
Recently, clinical trial data for these molecules designed for the treatment of ulcerative colitis and chronic lymphocytic leukemia, have demonstrated that parenterally or orally delivered oligonucleotides have specific therapeutic effect at their particular targets, providing more credibility to the treatment. The use of antisense oligonucleotide technologies in drug discovery is expected to address unmet medical needs. Therefore, the promise of antisense oligonucleotide technologies is stronger than ever.
Image Credit: Kateryna Kon/Shutterstock.com
References
- Aboul-Fadl, T., 2005. Antisense oligonucleotides: the state of the art. Current medicinal chemistry, 12(19), pp.2193-2214.
- Aboul-Fadl, T., 2006. Antisense oligonucleotide technologies in drug discovery. Expert Opinion on Drug Discovery, 1(4), pp.285-288.
- Behlke, M.A., 2006. Progress towards in vivo use of siRNAs. Molecular Therapy, 13(4), pp.644-670.
- Crooke, S.T., Liang, X.H., Crooke, R.M., Baker, B.F. and Geary, R.S., 2020. Antisense Drug Discovery and Development Technology Considered in a Pharmacological Context. Biochemical Pharmacology, p.114196.
- Tauchi, T., Sumi, M., Nakajima, A., Sashida, G., Shimamoto, T., and Ohyashiki, K., 2003. BCL-2 antisense oligonucleotide genasense is active against imatinib-resistant BCR-ABL-positive cells. Clinical cancer research, 9(11), pp.4267-4273.
- Van Aerschot, A., 2006. Oligonucleotides as antivirals: dream or realistic perspective?. Antiviral Research, 71(2-3), pp.307-316.
- Van Deventer, S.J.H., Tami, J.A. and Wedel, M.K., 2004. A randomized, controlled, double-blind, escalating dose study of alicaforsen enema in active ulcerative colitis. Gut, 53(11), pp.1646-1651.
- Yamamoto, T., Nakatani, M., Narukawa, K., and Obika, S., 2011. Antisense drug discovery and development. Future medicinal chemistry, 3(3), pp.339-365.
Further Reading
Last Updated: Nov 18, 2020