Antibodies, also known as immunoglobulins, are Y-shaped proteins produced by the immune system as a defense against infections. Each antibody targets a specific molecule present on the pathogen called antigens and binds to it with high specificity.
Image Credit: Lightspring/Shutterstock.com
The binding signals the immune system of potential infection and prevents the pathogen from entering host cells. However, pathogens such as viruses can have various types of mutations occurring at random that change the structure of the antigen thus, making the antigen unrecognizable by antibodies. These are known as antibody-escape mutant viruses.
How do antibodies work?
Antibodies are produced by immune cells called plasma cells as the first line of defense against infections. They are secreted into the bloodstream and the mucosa, where they recognize antigens and toxins, bind to and inactivate them. This process is known as neutralization.
Also, antigen-bound antibodies can recruit the complement system, which is an antibacterial protein system that forms pores on the surface of the pathogen, killing the pathogen. Moreover, pathogens bound with antibodies are signals for phagocytic cells to engulf the complex and eliminate foreign substances.
Evolution of antibody-escape mutant viruses
Many viruses evolve rapidly with numbers of mutations occurring randomly in the viral genome. Some mutations are silent, meaning the mutated gene encodes the same protein with the same amino acid composition and structure. However, sometimes mutations can be non-synonymous, meaning that the protein encoded would be altered and different from the non-mutated form.
When viruses are exposed to antibodies, there is a selection pressure against viruses that possess antigens recognized by the antibodies. As a result, these viruses are targeted by the antibodies and are killed. Nevertheless, for viral mutants that have mutations that change the antigen, they cannot be recognized by the antibodies. Thus, these viral mutants can survive and replicate.
Viruses can escape antibodies by single mutations. For example, the Influenza virus can escape most antibodies targeting the H1 haemagglutinin with a single mutation. However, there is always a cost to mutations. Although mutations can allow viruses to escape antibody selection, it may damage the functionality of the viral surface protein antigen.
The haemagglutinin protein of the Influenza virus is responsible for binding the virus to the infected cell. There are certain locations of the protein that is intolerable to mutations resulting in a huge change in structure as this renders the protein non-functional.
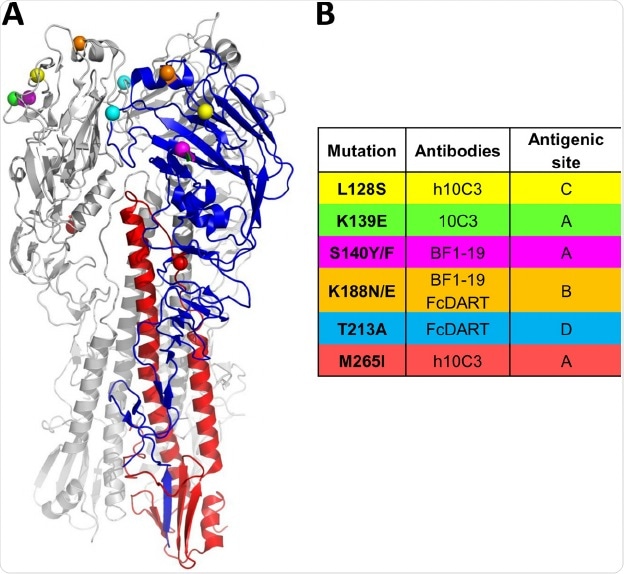
Image Credit: An Anti-H5N1 Influenza Virus FcDART Antibody Is a Highly Efficacious Therapeutic Agent and Prophylactic against H5N1 Influenza Virus Infection - Scientific Figure on ResearchGate. Available from: https://www.researchgate.net/figure/Monoclonal-antibody-escape-mutant-viruses-have-amino-acid-changes-located-primarily-in_fig4_272188296
Tackling antibody-escape mutant viruses
To eradicate viral diseases, researchers focus on designing antibodies that can effectively neutralize viruses and recognize escape mutants. For example, a chimeric human-murine antibody is recently designed and produced to capture multiple escape mutants of the human Hepatitis B virus. Alternatively, a cocktail of antibodies that targets different regions of the viral antigen can be given to patients to prevent the generation of antibody-escape mutant viruses.
Significance
Identifying viral escape from neutralizing antibodies is important for interpreting immune specificities and developing more effective therapeutic approaches to viral diseases. Furthermore, understanding viral antigenic evolution provides insight into vaccine design as vaccines such as the flu shot require the prediction of antigenic mutations of the Influenza virus to produce the vaccine in time.
References:
- Magnus, Carsten, Reh, Lucia, & Trkola, Alexandra. (2016). HIV-1 resistance to neutralizing antibodies: Determination of antibody concentrations leading to escape mutant evolution. Virus Research, 218, 57-70.
- Doud, Michael B, Hensley, Scott E, & Bloom, Jesse D. (2017). Complete mapping of viral escape from neutralizing antibodies. PLoS Pathogens, 13(3), E1006271.
- Doud, M. B., Lee, J. M., & Bloom, J. D. (2018). How single mutations affect viral escape from broad and narrow antibodies to H1 influenza hemagglutinin. Nature communications, 9(1), 1386. https://doi.org/10.1038/s41467-018-03665-3
- Baum, Alina, Fulton, Benjamin O, Wloga, Elzbieta, Copin, Richard, Pascal, Kristen E, Russo, Vincenzo, . . . Kyratsous, Christos A. (2020). Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science (American Association for the Advancement of Science), 369(6506), 1014.
- Golsaz-Shirazi, Forough, Amiri, Mohammad Mehdi, Farid, Samira, Bahadori, Motahareh, Bohne, Felix, Altstetter, Sebastian, . . . Shokri, Fazel. (2017). Construction of a hepatitis B virus-neutralizing chimeric monoclonal antibody recognizing escape mutants of the viral surface antigen (HBsAg). Antiviral Research, 144, 153-163.
Further Reading